Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution - PubMed (original) (raw)
Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution
James A Cahill et al. PLoS Genet. 2013.
Abstract
Despite extensive genetic analysis, the evolutionary relationship between polar bears (Ursus maritimus) and brown bears (U. arctos) remains unclear. The two most recent comprehensive reports indicate a recent divergence with little subsequent admixture or a much more ancient divergence followed by extensive admixture. At the center of this controversy are the Alaskan ABC Islands brown bears that show evidence of shared ancestry with polar bears. We present an analysis of genome-wide sequence data for seven polar bears, one ABC Islands brown bear, one mainland Alaskan brown bear, and a black bear (U. americanus), plus recently published datasets from other bears. Surprisingly, we find clear evidence for gene flow from polar bears into ABC Islands brown bears but no evidence of gene flow from brown bears into polar bears. Importantly, while polar bears contributed <1% of the autosomal genome of the ABC Islands brown bear, they contributed 6.5% of the X chromosome. The magnitude of sex-biased polar bear ancestry and the clear direction of gene flow suggest a model wherein the enigmatic ABC Island brown bears are the descendants of a polar bear population that was gradually converted into brown bears via male-dominated brown bear admixture. We present a model that reconciles heretofore conflicting genetic observations. We posit that the enigmatic ABC Islands brown bears derive from a population of polar bears likely stranded by the receding ice at the end of the last glacial period. Since then, male brown bear migration onto the island has gradually converted these bears into an admixed population whose phenotype and genotype are principally brown bear, except at mtDNA and X-linked loci. This process of genome erosion and conversion may be a common outcome when climate change or other forces cause a population to become isolated and then overrun by species with which it can hybridize.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
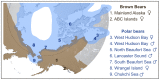
Figure 1. Map showing the approximate current geographic ranges of brown bears (brown) and polar bears (blue).
Numbers indicate the geographic location of origin of two brown bears and seven polar bears analyzed here. An American black bear from central Pennsylvania was also sequenced as part of this study. Shotgun data amounting to 4–6× coverage for polar bears and 11–12× coverage for brown and black bears (Table S2) was aligned to the current distribution of the polar bear genome .
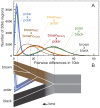
Figure 2. Genetic diversity within and between bear species.
(A) Pairwise differences between individuals estimated as the average number of differences per 10 thousand bases (kb) in 42,000 non-overlapping 50 kb regions. After strict quality filtering, within-sample heterozygosity was resolved by selecting a single, high-quality base at random. The Lancaster Sound polar bear showed an excess of postmortem damage, as expected for historic specimens , and is shown in Figure S1. Polar bears are remarkably homogenous compared to brown bears, and both polar bears and brown bears are approximately equally diverged from the American black bear. Consistent with the results of the _D-_statistic test, pairwise distance between the ABC Islands brown bear and all polar bears (yellow lines) is less than that between the mainland brown bear and all polar bears (red lines). (B) Schematic diagram of a representative gene tree within brown bear, polar bear, and black bear populations, with the present day at the left of the diagram. For this locus, admixture occurring more recently than the population divergence of polar bears leads to the introgression of a polar bear haplotype into brown bears. Estimate of average genomic distance for brown, black, and polar bears and for population divergence between brown bears and polar bears given different calibration points are provided in Table 1.
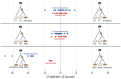
Figure 3. Summary of D-statistic comparisons between polar bears and brown bears.
In each comparison, the black bear was used to define the ancestral allele. The Z-score of the D-statistic for each comparison is shown for autosomes (red) and X-chromosome (blue). Each dot represents the data from comparison of one pair of bears. In the top panel, all pairs of polar bears are compared for excess derived allele matching against the mainland brown bear. In the middle panel, all pairs of polar bears are compared against the ABC Island brown bear. The bottom panel shows the comparison of the two brown bears for excess allele matching to polar bears with each dot representing a different polar bear.
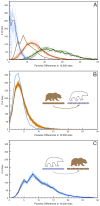
Figure 4. Simulated admixture reveals the direction of gene flow on the X chromosome.
(A) Pairwise distance as in Figure 2 but limited to the 12 scaffolds identified as X-chromosome. (B) 100 replicate simulations in which 6.5% of the female West Hudson Bay polar bear X-chromosome is replaced with that of the mainland Alaska brown bear in randomly inserted 20 kb fragments, simulating admixture from the brown bear genome into polar bear ∼50kya. Pairwise differences are calculated between the simulated genome (light brown lines; mean highlighted in dark brown) and the plot comparing the two female polar bears (blue line), to maximize the number of informative sites in the test. The addition of brown bear DNA to the polar bear genome markedly increases the number of high-diversity bins (>10 differences/10 kb), indicating that any introgression of brown bear DNA into polar bears should be easily detectable. (C). As in (B), but with 6.5% of the mainland Alaska brown bear X-chromosome is replaced with that of the female West Hudson Bay polar bear. In this instance, we find no difference between the simulated (blue lines) and real (brown line) data.
Similar articles
- Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears.
Cahill JA, Stirling I, Kistler L, Salamzade R, Ersmark E, Fulton TL, Stiller M, Green RE, Shapiro B. Cahill JA, et al. Mol Ecol. 2015 Mar;24(6):1205-17. doi: 10.1111/mec.13038. Epub 2015 Feb 5. Mol Ecol. 2015. PMID: 25490862 Free PMC article. - Introgressive hybridization: brown bears as vectors for polar bear alleles.
Hailer F. Hailer F. Mol Ecol. 2015 Mar;24(6):1161-3. doi: 10.1111/mec.13101. Mol Ecol. 2015. PMID: 25775930 - Genomic Evidence of Widespread Admixture from Polar Bears into Brown Bears during the Last Ice Age.
Cahill JA, Heintzman PD, Harris K, Teasdale MD, Kapp J, Soares AER, Stirling I, Bradley D, Edwards CJ, Graim K, Kisleika AA, Malev AV, Monaghan N, Green RE, Shapiro B. Cahill JA, et al. Mol Biol Evol. 2018 May 1;35(5):1120-1129. doi: 10.1093/molbev/msy018. Mol Biol Evol. 2018. PMID: 29471451 - New insights into dietary management of polar bears (Ursus maritimus) and brown bears (U. arctos).
Robbins CT, Tollefson TN, Rode KD, Erlenbach JA, Ardente AJ. Robbins CT, et al. Zoo Biol. 2022 Mar;41(2):166-175. doi: 10.1002/zoo.21658. Epub 2021 Nov 18. Zoo Biol. 2022. PMID: 34793606 Review. - Trichinella and polar bears: a limited risk for humans.
Dupouy-Camet J, Bourée P, Yera H. Dupouy-Camet J, et al. J Helminthol. 2017 Jul;91(4):440-446. doi: 10.1017/S0022149X17000219. Epub 2017 Apr 4. J Helminthol. 2017. PMID: 28372597 Review.
Cited by
- Analyses of key genes involved in Arctic adaptation in polar bears suggest selection on both standing variation and de novo mutations played an important role.
Samaniego Castruita JA, Westbury MV, Lorenzen ED. Samaniego Castruita JA, et al. BMC Genomics. 2020 Aug 6;21(1):543. doi: 10.1186/s12864-020-06940-0. BMC Genomics. 2020. PMID: 32758141 Free PMC article. - Discoveries and advances in plant and animal genomics.
Appels R, Nystrom J, Webster H, Keeble-Gagnere G. Appels R, et al. Funct Integr Genomics. 2015 Mar;15(2):121-9. doi: 10.1007/s10142-015-0434-3. Epub 2015 Mar 13. Funct Integr Genomics. 2015. PMID: 25763751 Free PMC article. Review. - Dental Calculus as a Tool to Study the Evolution of the Mammalian Oral Microbiome.
Brealey JC, Leitão HG, van der Valk T, Xu W, Bougiouri K, Dalén L, Guschanski K. Brealey JC, et al. Mol Biol Evol. 2020 Oct 1;37(10):3003-3022. doi: 10.1093/molbev/msaa135. Mol Biol Evol. 2020. PMID: 32467975 Free PMC article. - Does Effective Population Size Govern Evolutionary Differences in Telomere Length?
Brown LM, Elbon MC, Bharadwaj A, Damle G, Lachance J. Brown LM, et al. Genome Biol Evol. 2024 May 2;16(5):evae111. doi: 10.1093/gbe/evae111. Genome Biol Evol. 2024. PMID: 38771124 Free PMC article. - The origins and diversification of Holarctic brown bear populations inferred from genomes of past and present populations.
Segawa T, Rey-Iglesia A, Lorenzen ED, Westbury MV. Segawa T, et al. Proc Biol Sci. 2024 Jan 31;291(2015):20232411. doi: 10.1098/rspb.2023.2411. Epub 2024 Jan 24. Proc Biol Sci. 2024. PMID: 38264778
References
- Slater GJ, Figueirido B, Louis L, Yang P, Van Valkenburgh B (2010) Biomechanical consequences of rapid evolution in the polar bear lineage. PLoS ONE 5: e13870 doi:10.1371/journal.pone.0013870. - DOI - PMC - PubMed
- Stirling I (2011) Polar Bears: The Natural History of a Threatened Species. Brighton, Mass: Fitzhenry and Whiteside.
- Hailer F, Kutschera VE, Hallstrom BM, Klassert D, Fain SR, et al. (2012) Nuclear Genomic Sequences Reveal that Polar Bears Are an Old and Distinct Bear Lineage. Science 336: 344–347. - PubMed
- Kurtén B (1964) The evolution of the polar bear, Ursus maritimus (Phipps). Acta Zool Fenn 108: 1–26.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources