Reconstructing Roma history from genome-wide data - PubMed (original) (raw)
doi: 10.1371/journal.pone.0058633. Epub 2013 Mar 13.
Nick Patterson, Po-Ru Loh, Mark Lipson, Péter Kisfali, Bela I Melegh, Michael Bonin, Ludevít Kádaši, Olaf Rieß, Bonnie Berger, David Reich, Béla Melegh
Affiliations
- PMID: 23516520
- PMCID: PMC3596272
- DOI: 10.1371/journal.pone.0058633
Reconstructing Roma history from genome-wide data
Priya Moorjani et al. PLoS One. 2013.
Abstract
The Roma people, living throughout Europe and West Asia, are a diverse population linked by the Romani language and culture. Previous linguistic and genetic studies have suggested that the Roma migrated into Europe from South Asia about 1,000-1,500 years ago. Genetic inferences about Roma history have mostly focused on the Y chromosome and mitochondrial DNA. To explore what additional information can be learned from genome-wide data, we analyzed data from six Roma groups that we genotyped at hundreds of thousands of single nucleotide polymorphisms (SNPs). We estimate that the Roma harbor about 80% West Eurasian ancestry-derived from a combination of European and South Asian sources-and that the date of admixture of South Asian and European ancestry was about 850 years before present. We provide evidence for Eastern Europe being a major source of European ancestry, and North-west India being a major source of the South Asian ancestry in the Roma. By computing allele sharing as a measure of linkage disequilibrium, we estimate that the migration of Roma out of the Indian subcontinent was accompanied by a severe founder event, which appears to have been followed by a major demographic expansion after the arrival in Europe.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
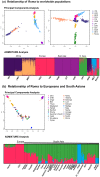
Figure 1. Relationship of Roma with other worldwide populations.
We applied PCA and ADMIXTURE to study the relationship of Roma with the HapMap and South Asian populations. In PCA, each point represents an individual, and in ADMIXTURE, each line represents an individual. (a) shows the PCA and ADMIXTURE results for clustering of Roma and HapMap populations. The populations codes are as follows: Yoruba in Ibadan, Nigeria (YRI), Luhya in Webuye, Kenya (LWK), Maasai in Kinyawa, Kenya (MKK), Utah residents with Northern and Western European ancestry (CEU), Toscani in Italia (TSI), Han Chinese in Beijing, China (CHB), Japanese in Tokyo, Japan (JPT), Chinese in Metropolitan Denver, Colorado (CHD), Gujarati Indians in Houston, Texas (GIH), African ancestry in Southwest USA (ASW) and Mexican ancestry in Los Angeles, California (MEX), and (b) shows the PCA and ADMIXTURE results for clustering of Roma and South Asian groups. We limit the sample size of all groups (except Roma) to 20 individuals.
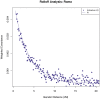
Figure 2. Admixture date estimation.
We performed ROLLOFF (using R(d)) on the Roma samples (n = 24). We plot the weighted covariance as a function of genetic distance, and obtain a date by fitting an exponential function with an affine term:  , where d is the genetic distance in Morgans and n is the number of generations since mixture. We do not show inter-SNP intervals of <0.5 cM since we have found that at this distance admixture LD begins to be confounded by background LD.
, where d is the genetic distance in Morgans and n is the number of generations since mixture. We do not show inter-SNP intervals of <0.5 cM since we have found that at this distance admixture LD begins to be confounded by background LD.
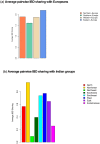
Figure 3. The European and South Asian sources of Roma ancestry.
We computed a genome-wide average IBD sharing distance between Roma (all samples combined in one group) and other regional groups. Details of the regional grouping are described in Methods. (a) shows the average pairwise IBD sharing between Roma and Europeans (grouped into four regional categories), (b) shows IBD sharing average pairwise IBD sharing between Roma and South Asians (grouped into 8 regional categories).
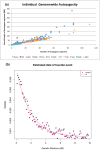
Figure 4. Founder events in the Roma.
(a) shows estimates of genomewide autozygosity in Roma and individuals from HapMap (n = 30 from each of the 11 HapMap populations). Each point represents an individual with the color-coding described in the legend. (b) shows the decay of autocorrelation with genetic distance. We fit an exponential function:  where D = distance in Morgans and t = time of founder event. We thus infer a founder event date of 27 generations.
where D = distance in Morgans and t = time of founder event. We thus infer a founder event date of 27 generations.
Similar articles
- European Roma groups show complex West Eurasian admixture footprints and a common South Asian genetic origin.
Font-Porterias N, Arauna LR, Poveda A, Bianco E, Rebato E, Prata MJ, Calafell F, Comas D. Font-Porterias N, et al. PLoS Genet. 2019 Sep 23;15(9):e1008417. doi: 10.1371/journal.pgen.1008417. eCollection 2019 Sep. PLoS Genet. 2019. PMID: 31545809 Free PMC article. - Sex-biased patterns shaped the genetic history of Roma.
García-Fernández C, Font-Porterias N, Kučinskas V, Sukarova-Stefanovska E, Pamjav H, Makukh H, Dobon B, Bertranpetit J, Netea MG, Calafell F, Comas D. García-Fernández C, et al. Sci Rep. 2020 Sep 2;10(1):14464. doi: 10.1038/s41598-020-71066-y. Sci Rep. 2020. PMID: 32879340 Free PMC article. - Refining the South Asian Origin of the Romani people.
Melegh BI, Banfai Z, Hadzsiev K, Miseta A, Melegh B. Melegh BI, et al. BMC Genet. 2017 Aug 31;18(1):82. doi: 10.1186/s12863-017-0547-x. BMC Genet. 2017. PMID: 28859608 Free PMC article. - Revealing the impact of the Caucasus region on the genetic legacy of Romani people from genome-wide data.
Bánfai Z, Ádám V, Pöstyéni E, Büki G, Czakó M, Miseta A, Melegh B. Bánfai Z, et al. PLoS One. 2018 Sep 10;13(9):e0202890. doi: 10.1371/journal.pone.0202890. eCollection 2018. PLoS One. 2018. PMID: 30199533 Free PMC article. - Origin of ethnic groups, linguistic families, and civilizations in China viewed from the Y chromosome.
Yu X, Li H. Yu X, et al. Mol Genet Genomics. 2021 Jul;296(4):783-797. doi: 10.1007/s00438-021-01794-x. Epub 2021 May 26. Mol Genet Genomics. 2021. PMID: 34037863 Review.
Cited by
- Analysis of Gyimes Csango population samples on a high-resolution genome-wide basis.
Bánfai Z, Büki G, Ádám V, Sümegi K, Szabó A, Hadzsiev K, Erős K, Gallyas F, Miseta A, Kásler M, Melegh B. Bánfai Z, et al. BMC Genomics. 2024 Oct 7;25(1):942. doi: 10.1186/s12864-024-10833-x. BMC Genomics. 2024. PMID: 39375616 Free PMC article. - Population history modulates the fitness effects of Copy Number Variation in the Roma.
Antinucci M, Comas D, Calafell F. Antinucci M, et al. Hum Genet. 2023 Sep;142(9):1327-1343. doi: 10.1007/s00439-023-02579-5. Epub 2023 Jun 14. Hum Genet. 2023. PMID: 37311904 Free PMC article. - Methods for Assessing Population Relationships and History Using Genomic Data.
Moorjani P, Hellenthal G. Moorjani P, et al. Annu Rev Genomics Hum Genet. 2023 Aug 25;24:305-332. doi: 10.1146/annurev-genom-111422-025117. Epub 2023 May 23. Annu Rev Genomics Hum Genet. 2023. PMID: 37220313 Free PMC article. Review. - Population Genetics of the European Roma-A Review.
Ena GF, Aizpurua-Iraola J, Font-Porterias N, Calafell F, Comas D. Ena GF, et al. Genes (Basel). 2022 Nov 8;13(11):2068. doi: 10.3390/genes13112068. Genes (Basel). 2022. PMID: 36360305 Free PMC article. Review. - Founder lineages in the Iberian Roma mitogenomes recapitulate the Roma diaspora and show the effects of demographic bottlenecks.
Aizpurua-Iraola J, Giménez A, Carballo-Mesa A, Calafell F, Comas D. Aizpurua-Iraola J, et al. Sci Rep. 2022 Nov 4;12(1):18720. doi: 10.1038/s41598-022-23349-9. Sci Rep. 2022. PMID: 36333436 Free PMC article.
References
- Liégeois JP (1994) Roma, gypsies, travellers: Sales Agent Manhattan Pub. Co. Distributor.
- Marushiakova E, Popov V (1997) Gypsies (Roma) in Bulgaria: P. Lang.
- Fraser AM (1995) The gypsies: Wiley-Blackwell.
- Kalaydjieva L, Morar B, Chaix R, Tang H (2005) A newly discovered founder population: the Roma/Gypsies. Bioessays 27: 1084–1094. - PubMed
- Schurr TG (2004) Reconstructing the origins and migrations of diasporic populations: the case of the European Gypsies. American anthropologist 106: 267–281.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources