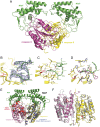The E. coli effector protein NleF is a caspase inhibitor - PubMed (original) (raw)
doi: 10.1371/journal.pone.0058937. Epub 2013 Mar 14.
Mario Mörtl, Holger Steuber, Gabriella Siszler, Shahista Nisa, Frank Schwarz, Inna Lavrik, Thomas M A Gronewold, Klaus Maskos, Michael S Donnenberg, Dirk Ullmann, Peter Uetz, Manfred Kögl
Affiliations
- PMID: 23516580
- PMCID: PMC3597564
- DOI: 10.1371/journal.pone.0058937
The E. coli effector protein NleF is a caspase inhibitor
Sonja Blasche et al. PLoS One. 2013.
Abstract
Enterohemorrhagic and enteropathogenic E. coli (EHEC and EPEC) can cause severe and potentially life-threatening infections. Their pathogenicity is mediated by at least 40 effector proteins which they inject into their host cells by a type-III secretion system leading to the subversion of several cellular pathways. However, the molecular function of several effectors remains unknown, even though they contribute to virulence. Here we show that one of them, NleF, binds to caspase-4, -8, and -9 in yeast two-hybrid, LUMIER, and direct interaction assays. NleF inhibits the catalytic activity of the caspases in vitro and in cell lysate and prevents apoptosis in HeLa and Caco-2 cells. We have solved the crystal structure of the caspase-9/NleF complex which shows that NleF uses a novel mode of caspase inhibition, involving the insertion of the carboxy-terminus of NleF into the active site of the protease. In conformance with our structural model, mutagenized NleF with truncated or elongated carboxy-termini revealed a complete loss in caspase binding and apoptosis inhibition. Evasion of apoptosis helps pathogenic E. coli and other pathogens to take over the host cell by counteracting the cell's ability to self-destruct upon infection. Recently, two other effector proteins, namely NleD and NleH, were shown to interfere with apoptosis. Even though NleF is not the only effector protein capable of apoptosis inhibition, direct inhibition of caspases by bacterial effectors has not been reported to date. Also unique so far is its mode of inhibition that resembles the one obtained for synthetic peptide-type inhibitors and as such deviates substantially from previously reported caspase-9 inhibitors such as the BIR3 domain of XIAP.
Conflict of interest statement
Competing Interests: Dr. Gronewold is an employee of SAW. Mario Mörtl and Klaus Maskos are affiliated with Proteros. Dirk Ullmann and Holger Steuber used to work at Proteros, the company that solved the crystal structure of the complex we describe. This does not alter the authors’ adherence to all the PLOS ONE policies on sharing data and materials. All other authors, including Dirk Ullmann and Holger Steuber, declare no competing interest.
Figures
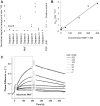
Figure 1. NleF binds caspases -4, -8 and -9.
A. Binding of NleF to caspases-4, -8 and -9 using luciferase-NleF and protein-A-caspase fusions (LUMIER assays34). Interaction strengths are expressed as signal to background ratios using binding to protein A as a negative control. The interaction of JUN to FOS serves as a positive control. Squares indicate individual measurements. B,C. The affinity constant (Kd) of NleF binding to immobilized caspase-9 as measured by surface acoustic wave (SAW) technology is ∼39 nM. The Kd is defined as follows: Kd = Koff/Kon = y-intercept/slope (C), SAW overlay plot of NleF injections. NleF concentrations are indicated in the legend. The two bold curves added to each NleF measurement (on the left and right of the peak) represent the optimal Kobs and Koff curves.
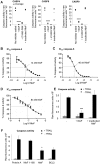
Figure 2. NleF inhibits caspases in vitro and in in cells.
A. In vitro inhibition of caspase-4 (1 U), caspase-8 (1 U) and caspase-9 (1 U) by NleF (1.5 µg) (1.5 µg/unit caspase), the inhibitor Z-VAD-fluoromethylketone (Z-VAD-FMK, 20 µM), and proteinase K-inactivated NleF (1.5 µg). B-D. Dose-dependent inhibition of purified caspase-4, -8, and -9 by NleF (logarithmic concentration indicated in the X axis): B. Caspase-4 (200 nM). C. caspase-8 (68 nM). D. caspase-9 (1.1 µM). E. Recombinant NleF inhibits caspase-9 activity in lysates of apoptotic cells. HeLa cells were induced to enter apoptosis by treatment with TRAIL (25 ng/ml) for four hours (black bars) or left untreated (striped bars) before preparation of lysates and measurement of cellular caspase-9 activity in the absence or presence of an abundant amount of purified NleF. F. Caspase-9 activity in apoptotic HeLa cell extracts expressing Protein A, an inactive fragment of NleF (amino acids 1–160), wild type NleF or BCL2, respectively. Significance was determined using the two-tailed unpaired Student’s t-test. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
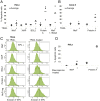
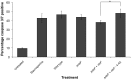
Figure 3. NleF inhibits apoptosis induced by TRAIL and staurosporine.
A–C. Percentage of apoptotic HeLa and Caco-2 cells in TRAIL (25 ng/ml) treated (+) and untreated (−) samples. A. HeLa cells expressing YFP-fusions of wild type NleF, XIAP and BCL2, respectively, exhibited decreased apoptosis in comparison to cells expressing Protein A or an inactive NleF fragment (amino acids 1–160) after TRAIL treatment. Squares: percentage of apoptotic cells (three independent experiments); bars: average. B. Caco-2 cells expressing NleF and Protein A, respectively. Significance (t-test): *P<0.05. C. Representative FACS counts of HeLa cells expressing the indicated YFP-tagged constructs. Cells that stained positive for annexin V- allophycocyanine (APC+) but negative for propidium iodide were counted as apoptotic cells. D. Percentage of apoptotic HeLa cells in staurosporine treated (+) and untreated (−) samples expressing NleF and Protein A, respectively.
Figure 4. Crystal structure of Caspase-9 with bound NleF.
A. The caspase-9 protease monomers are shown in magenta and yellow, the active site cysteines (Cys285) are shown as red sticks for improved special orientation of the catalytic site. The NleF effector proteins attached to the caspase active sites are shown in green, as labeled in the figure. B. The C-terminal residues of NleF are anchoring the effector protein to the specificity pockets of the protease. The Fo-Fc omit electron density contoured at 2.5 σ is shown in blue for a representative part of the involved C-terminal NleF residues. C. Detailed binding mode of the NleF C-terminus to the caspase-9 active site. Involved amino acids are shown as green and yellow sticks, hydrogen bonds are depicted as magenta dashed lines. D. Superposition of the NleF(green)-bound caspase-9 (yellow) with the peptide inhibitor Z-EVD-Dcbmk (magenta) bound to the caspase-9 monomer (blue) captured in active state (PDB entry 1JXQ). While the conformation of caspase residues is virtually identical, slight differences in the mode of interaction of the bound inhibitors are e.g. observed for the Asp/Gly-P1 moiety, where the Asp of Z-EVD-Dcbmk adopts a more favourable geometry for efficient salt bridge formation to Arg179. E. Superimposition of the NleF-bound caspase-9 dimer presented in this study and the previously described caspase-9 crystal structure (light blue) in complex with the BIR3 domain of the XIAP inhibitor shown as red cartoon (PDB entry 1NW9). While the latter interferes with the formation of a productive caspase dimer, NleF establishes its inhibitory activity by means of a fundamentally different mechanism of active site targeting. F. Superimposition of the caspase-9 dimer obtained for the NleF-inhibited form (yellow and magenta as in A., both NleF molecules omitted for clarity) and the dimer of 1JXQ (grey cartoon). While the type of dimer formation and the overall protein conformation is well conserved between the two dimers, deviations arise mainly from the fact that one of the two 1JXQ monomers (left) corresponds to a non-productive, inactive conformation while in the NleF-inhibited dimer, both monomers adopt an active state with well-formed specificity pockets.
Figure 5. Caspase 3/7 activity in infected HeLa cells.
HeLa cells were infected with wild type EPEC strain E2348/69, Δ_nle_F deletion mutant, or Δ_nle_F complemented in single copy with _nle_F variants for 2.5 to 3 hours. Treatment of HeLa cells with staurosporine (5 µM) was used as a positive control. There was no significant difference between caspase activity upon infection with the wild type strain and the Δ_nle_F deletion mutant, however the deletion mutant complemented with the wild type allele consistently induced significantly less caspase 3/7 activity than the deletion mutant complemented with the allele lacking the last 4 codons. Significance was determined using the two-tailed Wilcoxon test *p<0.02.
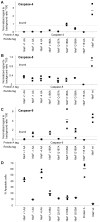
Figure 6. Caspase binding and apoptosis inhibition by modified versions of NleF.
A–C. Caspase-9-binding of different versions of NleF assessed by LUMIER assays. A. Caspase-4, B. caspase-8, C. caspase-9. D. Percentage of apoptotic HeLa cells expressing different versions of NleF after induction with staurosporine. NleF +1: NleF with an additional C-terminal alanine; NleF -1 and NleF -4: NleF with the terminal 1 and 4 amino acids removed, respectively; NleF L186A, NleF Q187A, NleF C188A and NleF G189A: NleF versions with indicated amino acids substituted by alanine; NleF +18: NleF with additional 18 amino acids.
Similar articles
- Distinct Roles of the Antiapoptotic Effectors NleB and NleF from Enteropathogenic Escherichia coli.
Pollock GL, Oates CVL, Giogha C, Wong Fok Lung T, Ong SY, Pearson JS, Hartland EL. Pollock GL, et al. Infect Immun. 2017 Mar 23;85(4):e01071-16. doi: 10.1128/IAI.01071-16. Print 2017 Apr. Infect Immun. 2017. PMID: 28138023 Free PMC article. - The type III secretion effector NleF of enteropathogenic Escherichia coli activates NF-κB early during infection.
Pallett MA, Berger CN, Pearson JS, Hartland EL, Frankel G. Pallett MA, et al. Infect Immun. 2014 Nov;82(11):4878-88. doi: 10.1128/IAI.02131-14. Epub 2014 Sep 2. Infect Immun. 2014. PMID: 25183730 Free PMC article. - A Type III Effector NleF from EHEC Inhibits Epithelial Inflammatory Cell Death by Targeting Caspase-4.
Song T, Li K, Zhou W, Zhou J, Jin Y, Dai H, Xu T, Hu M, Ren H, Yue J, Liang L. Song T, et al. Biomed Res Int. 2017;2017:4101745. doi: 10.1155/2017/4101745. Epub 2017 May 16. Biomed Res Int. 2017. PMID: 28593173 Free PMC article. - Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements.
Wong AR, Pearson JS, Bright MD, Munera D, Robinson KS, Lee SF, Frankel G, Hartland EL. Wong AR, et al. Mol Microbiol. 2011 Jun;80(6):1420-38. doi: 10.1111/j.1365-2958.2011.07661.x. Epub 2011 May 5. Mol Microbiol. 2011. PMID: 21488979 Review. - Bringing down the host: enteropathogenic and enterohaemorrhagic Escherichia coli effector-mediated subversion of host innate immune pathways.
Santos AS, Finlay BB. Santos AS, et al. Cell Microbiol. 2015 Mar;17(3):318-32. doi: 10.1111/cmi.12412. Epub 2015 Feb 4. Cell Microbiol. 2015. PMID: 25588886 Review.
Cited by
- Bacterial secreted effectors and caspase-3 interactions.
Wall DM, McCormick BA. Wall DM, et al. Cell Microbiol. 2014 Dec;16(12):1746-56. doi: 10.1111/cmi.12368. Epub 2014 Oct 30. Cell Microbiol. 2014. PMID: 25262664 Free PMC article. Review. - Caspase-4 and -5 Biology in the Pathogenesis of Inflammatory Bowel Disease.
Smith AP, Creagh EM. Smith AP, et al. Front Pharmacol. 2022 May 31;13:919567. doi: 10.3389/fphar.2022.919567. eCollection 2022. Front Pharmacol. 2022. PMID: 35712726 Free PMC article. Review. - The EHEC-host interactome reveals novel targets for the translocated intimin receptor.
Blasche S, Arens S, Ceol A, Siszler G, Schmidt MA, Häuser R, Schwarz F, Wuchty S, Aloy P, Uetz P, Stradal T, Koegl M. Blasche S, et al. Sci Rep. 2014 Dec 18;4:7531. doi: 10.1038/srep07531. Sci Rep. 2014. PMID: 25519916 Free PMC article. - Mechanisms Applied by Protein Inhibitors to Inhibit Cysteine Proteases.
Tušar L, Usenik A, Turk B, Turk D. Tušar L, et al. Int J Mol Sci. 2021 Jan 20;22(3):997. doi: 10.3390/ijms22030997. Int J Mol Sci. 2021. PMID: 33498210 Free PMC article. Review. - Diversity, Distribution and Structural Prediction of the Pathogenic Bacterial Effectors EspN and EspS.
Li Z, Hu Y, Song Y, Li D, Yang X, Zhang L, Li T, Wang H. Li Z, et al. Genes (Basel). 2024 Sep 26;15(10):1250. doi: 10.3390/genes15101250. Genes (Basel). 2024. PMID: 39457374 Free PMC article.
References
- CDC (2006) Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach–United States, September 2006. MMWR Morb Mortal Wkly Rep 55: 1045–1046. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases