A revised timescale for human evolution based on ancient mitochondrial genomes - PubMed (original) (raw)
. 2013 Apr 8;23(7):553-559.
doi: 10.1016/j.cub.2013.02.044. Epub 2013 Mar 21.
Alissa Mittnik # 3, Philip L F Johnson 4, Kirsten Bos 3 5, Martina Lari 6, Ruth Bollongino 7, Chengkai Sun 8, Liane Giemsch 9 10, Ralf Schmitz 9, Joachim Burger 7, Anna Maria Ronchitelli 11, Fabio Martini 12, Renata G Cremonesi 13, Jiří Svoboda 14 15, Peter Bauer 16, David Caramelli 6, Sergi Castellano 1, David Reich 17 18, Svante Pääbo 1, Johannes Krause 3
Affiliations
- PMID: 23523248
- PMCID: PMC5036973
- DOI: 10.1016/j.cub.2013.02.044
A revised timescale for human evolution based on ancient mitochondrial genomes
Qiaomei Fu et al. Curr Biol. 2013.
Abstract
Background: Recent analyses of de novo DNA mutations in modern humans have suggested a nuclear substitution rate that is approximately half that of previous estimates based on fossil calibration. This result has led to suggestions that major events in human evolution occurred far earlier than previously thought.
Results: Here, we use mitochondrial genome sequences from ten securely dated ancient modern humans spanning 40,000 years as calibration points for the mitochondrial clock, thus yielding a direct estimate of the mitochondrial substitution rate. Our clock yields mitochondrial divergence times that are in agreement with earlier estimates based on calibration points derived from either fossils or archaeological material. In particular, our results imply a separation of non-Africans from the most closely related sub-Saharan African mitochondrial DNAs (haplogroup L3) that occurred less than 62-95 kya.
Conclusions: Though single loci like mitochondrial DNA (mtDNA) can only provide biased estimates of population divergence times, they can provide valid upper bounds. Our results exclude most of the older dates for African and non-African population divergences recently suggested by de novo mutation rate estimates in the nuclear genome.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
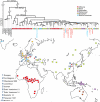
Figure 1. Tree for 54 present-day humans, 10 ancient modern humans, and 7 archaic humans
The phylogeny in the top panel was constructed using Maximum Parsimony and rooted using midpoint rooting. The branches for present-day humans do not all end at the same point giving a sense of the inherent uncertainty in time measurements based on mtDNA due to its limited sequence span. However, the consistent shortening of the branches of ancient humans relative to their closest present-day human relatives is apparent in the figure. This is the basis for our clock calibration. Pre- and post-Neolithic ancient samples are indicated as red and blue circles, respectively, and colored squares indicate the geographical origin of 54 present-day humans that we co-analyzed with them. Date estimates for major divergence events are shown at the nodes. In the bottom panel we show a map giving geographical origin of the samples.
Similar articles
- Improved calibration of the human mitochondrial clock using ancient genomes.
Rieux A, Eriksson A, Li M, Sobkowiak B, Weinert LA, Warmuth V, Ruiz-Linares A, Manica A, Balloux F. Rieux A, et al. Mol Biol Evol. 2014 Oct;31(10):2780-92. doi: 10.1093/molbev/msu222. Epub 2014 Aug 5. Mol Biol Evol. 2014. PMID: 25100861 Free PMC article. - Variation in the Substitution Rates among the Human Mitochondrial Haplogroup U Sublineages.
Översti S, Palo JU. Översti S, et al. Genome Biol Evol. 2022 Jul 2;14(7):evac097. doi: 10.1093/gbe/evac097. Genome Biol Evol. 2022. PMID: 35731946 Free PMC article. - Neolithic mitochondrial haplogroup H genomes and the genetic origins of Europeans.
Brotherton P, Haak W, Templeton J, Brandt G, Soubrier J, Jane Adler C, Richards SM, Der Sarkissian C, Ganslmeier R, Friederich S, Dresely V, van Oven M, Kenyon R, Van der Hoek MB, Korlach J, Luong K, Ho SYW, Quintana-Murci L, Behar DM, Meller H, Alt KW, Cooper A; Genographic Consortium. Brotherton P, et al. Nat Commun. 2013;4:1764. doi: 10.1038/ncomms2656. Nat Commun. 2013. PMID: 23612305 Free PMC article. - Archaic human genomics.
Disotell TR. Disotell TR. Am J Phys Anthropol. 2012;149 Suppl 55:24-39. doi: 10.1002/ajpa.22159. Epub 2012 Nov 2. Am J Phys Anthropol. 2012. PMID: 23124308 Review. - Catarrhine primate divergence dates estimated from complete mitochondrial genomes: concordance with fossil and nuclear DNA evidence.
Raaum RL, Sterner KN, Noviello CM, Stewart CB, Disotell TR. Raaum RL, et al. J Hum Evol. 2005 Mar;48(3):237-57. doi: 10.1016/j.jhevol.2004.11.007. Epub 2005 Jan 20. J Hum Evol. 2005. PMID: 15737392 Review.
Cited by
- Ancient genomes from the Tang Dynasty capital reveal the genetic legacy of trans-Eurasian communication at the eastern end of Silk Road.
Lv M, Ma H, Wang R, Li H, Zhang X, Zhang W, Zeng Y, Qin Z, Zhai H, Lou Y, Lin Y, Tao L, He H, Yang X, Zhu K, Zhou Y, Wang CC. Lv M, et al. BMC Biol. 2024 Nov 20;22(1):267. doi: 10.1186/s12915-024-02068-9. BMC Biol. 2024. PMID: 39567925 Free PMC article. - Leveraging ancient DNA to uncover signals of natural selection in Europe lost due to admixture or drift.
Pandey D, Harris M, Garud NR, Narasimhan VM. Pandey D, et al. Nat Commun. 2024 Nov 12;15(1):9772. doi: 10.1038/s41467-024-53852-8. Nat Commun. 2024. PMID: 39532856 Free PMC article. - The rise and transformation of Bronze Age pastoralists in the Caucasus.
Ghalichi A, Reinhold S, Rohrlach AB, Kalmykov AA, Childebayeva A, Yu H, Aron F, Semerau L, Bastert-Lamprichs K, Belinskiy AB, Berezina NY, Berezin YB, Broomandkhoshbacht N, Buzhilova AP, Erlikh VR, Fehren-Schmitz L, Gambashidze I, Kantorovich AR, Kolesnichenko KB, Lordkipanidze D, Magomedov RG, Malek-Custodis K, Mariaschk D, Maslov VE, Mkrtchyan L, Nagler A, Fazeli Nashli H, Ochir M, Piotrovskiy YY, Saribekyan M, Sheremetev AG, Stöllner T, Thomalsky J, Vardanyan B, Posth C, Krause J, Warinner C, Hansen S, Haak W. Ghalichi A, et al. Nature. 2024 Nov;635(8040):917-925. doi: 10.1038/s41586-024-08113-5. Epub 2024 Oct 30. Nature. 2024. PMID: 39478221 Free PMC article. - Life history and ancestry of the late Upper Palaeolithic infant from Grotta delle Mura, Italy.
Higgins OA, Modi A, Cannariato C, Diroma MA, Lugli F, Ricci S, Zaro V, Vai S, Vazzana A, Romandini M, Yu H, Boschin F, Magnone L, Rossini M, Di Domenico G, Baruffaldi F, Oxilia G, Bortolini E, Dellù E, Moroni A, Ronchitelli A, Talamo S, Müller W, Calattini M, Nava A, Posth C, Lari M, Bondioli L, Benazzi S, Caramelli D. Higgins OA, et al. Nat Commun. 2024 Sep 20;15(1):8248. doi: 10.1038/s41467-024-51150-x. Nat Commun. 2024. PMID: 39304646 Free PMC article. - Low Genetic Impact of the Roman Occupation of Britain in Rural Communities.
Scheib CL, Hui R, Rose AK, D'Atanasio E, Inskip SA, Dittmar J, Cessford C, Griffith SJ, Solnik A, Wiseman R, Neil B, Biers T, Harknett SJ, Sasso S, Biagini SA, Runfeldt G, Duhig C, Evans C, Metspalu M, Millett MJ, O'Connell TC, Robb JE, Kivisild T. Scheib CL, et al. Mol Biol Evol. 2024 Sep 4;41(9):msae168. doi: 10.1093/molbev/msae168. Mol Biol Evol. 2024. PMID: 39268685 Free PMC article.
References
- Scally A, Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–753. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources