Robust data-driven incorporation of prior knowledge into the inference of dynamic regulatory networks - PubMed (original) (raw)
Robust data-driven incorporation of prior knowledge into the inference of dynamic regulatory networks
Alex Greenfield et al. Bioinformatics. 2013.
Abstract
Motivation: Inferring global regulatory networks (GRNs) from genome-wide data is a computational challenge central to the field of systems biology. Although the primary data currently used to infer GRNs consist of gene expression and proteomics measurements, there is a growing abundance of alternate data types that can reveal regulatory interactions, e.g. ChIP-Chip, literature-derived interactions, protein-protein interactions. GRN inference requires the development of integrative methods capable of using these alternate data as priors on the GRN structure. Each source of structure priors has its unique biases and inherent potential errors; thus, GRN methods using these data must be robust to noisy inputs.
Results: We developed two methods for incorporating structure priors into GRN inference. Both methods [Modified Elastic Net (MEN) and Bayesian Best Subset Regression (BBSR)] extend the previously described Inferelator framework, enabling the use of prior information. We test our methods on one synthetic and two bacterial datasets, and show that both MEN and BBSR infer accurate GRNs even when the structure prior used has significant amounts of error (>90% erroneous interactions). We find that BBSR outperforms MEN at inferring GRNs from expression data and noisy structure priors.
Availability and implementation: Code, datasets and networks presented in this article are available at http://bonneaulab.bio.nyu.edu/software.html.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
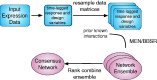
Fig. 1.
Method flow chart. Our method takes as input an expression dataset. To build a mechanistic model of gene expression, we create time-lagged response and design variables, such that the expression of the TF is time-lagged with respect to the expression of the target. We then resample the response and designing matrices, running model selection (using either
MEN
or
BBSR
) for each resample. This generates an ensemble of networks, which we rank combine into one final network
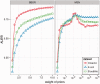
Fig. 2.
Effect of weight parameter on performance. We use all GSIs as the set of PKIs, and evaluate performance (in terms of AUPR) against the set of GSIs. We evaluate this performance for a variety of choices of the weight parameter for both methods
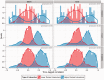
Fig. 3.
Incorporation of prior interactions is data driven. For all three datasets, we used all GSIs as PKIs. Here, we display the distribution of time-lagged correlation of predicted TF-target pairs at a recall level of  (higher ranked, blue), and low ranked interactions that are in the gold standard (lower ranked, red). Note that high ranked interactions are less likely to have low absolute time-lagged correlation, and the low ranked GSIs are centred around 0
(higher ranked, blue), and low ranked interactions that are in the gold standard (lower ranked, red). Note that high ranked interactions are less likely to have low absolute time-lagged correlation, and the low ranked GSIs are centred around 0
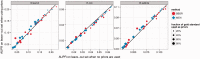
Fig. 4.
Performance change on the leave-out set. PKIs were sampled randomly from 20%, 40%, 60% and 80% of the GSIs in five repetitions. We define the leave-out set as the set of GSIs that are not PKIs. Here, we compare the AUPR of the leave-out set when using PKIs (_y_-axis) to the AUPR when not using PKIs (_x_-axis). Points above the line indicate a performance increase when PKIs are used
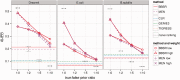
Fig. 5.
Robustness to incorrect prior information. For each dataset, we considered half of the GSIs as TPIs, and added varying numbers of FPIs that were not GSIs. We show the AUPR of both methods for multiple choices of the respective weight parameters, as well as methods that do not use any PKIs (horizontal lines). Additionally, we show the performance of a naive interaction ranking method, which places all PKIs at the top of the list (gray bars)
Similar articles
- Inference of gene networks from gene expression time series using recurrent neural networks and sparse MAP estimation.
Chen CK. Chen CK. J Bioinform Comput Biol. 2018 Aug;16(4):1850009. doi: 10.1142/S0219720018500099. Epub 2018 Apr 26. J Bioinform Comput Biol. 2018. PMID: 30051742 - MICRAT: a novel algorithm for inferring gene regulatory networks using time series gene expression data.
Yang B, Xu Y, Maxwell A, Koh W, Gong P, Zhang C. Yang B, et al. BMC Syst Biol. 2018 Dec 14;12(Suppl 7):115. doi: 10.1186/s12918-018-0635-1. BMC Syst Biol. 2018. PMID: 30547796 Free PMC article. - Inference of Gene Regulatory Network Based on Local Bayesian Networks.
Liu F, Zhang SW, Guo WF, Wei ZG, Chen L. Liu F, et al. PLoS Comput Biol. 2016 Aug 1;12(8):e1005024. doi: 10.1371/journal.pcbi.1005024. eCollection 2016 Aug. PLoS Comput Biol. 2016. PMID: 27479082 Free PMC article. - Gene regulatory network inference resources: A practical overview.
Mercatelli D, Scalambra L, Triboli L, Ray F, Giorgi FM. Mercatelli D, et al. Biochim Biophys Acta Gene Regul Mech. 2020 Jun;1863(6):194430. doi: 10.1016/j.bbagrm.2019.194430. Epub 2019 Oct 31. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 31678629 Review. - Computational prediction of gene regulatory networks in plant growth and development.
Haque S, Ahmad JS, Clark NM, Williams CM, Sozzani R. Haque S, et al. Curr Opin Plant Biol. 2019 Feb;47:96-105. doi: 10.1016/j.pbi.2018.10.005. Epub 2018 Nov 14. Curr Opin Plant Biol. 2019. PMID: 30445315 Review.
Cited by
- Network inference with Granger causality ensembles on single-cell transcriptomics.
Deshpande A, Chu LF, Stewart R, Gitter A. Deshpande A, et al. Cell Rep. 2022 Feb 8;38(6):110333. doi: 10.1016/j.celrep.2022.110333. Cell Rep. 2022. PMID: 35139376 Free PMC article. - High-Dimensional Bayesian Network Inference From Systems Genetics Data Using Genetic Node Ordering.
Wang L, Audenaert P, Michoel T. Wang L, et al. Front Genet. 2019 Dec 20;10:1196. doi: 10.3389/fgene.2019.01196. eCollection 2019. Front Genet. 2019. PMID: 31921278 Free PMC article. - dynGENIE3: dynamical GENIE3 for the inference of gene networks from time series expression data.
Huynh-Thu VA, Geurts P. Huynh-Thu VA, et al. Sci Rep. 2018 Feb 21;8(1):3384. doi: 10.1038/s41598-018-21715-0. Sci Rep. 2018. PMID: 29467401 Free PMC article. - Benchmarking of protein interaction databases for integration with manually reconstructed signalling network models.
Van de Graaf MW, Eggertsen TG, Zeigler AC, Tan PM, Saucerman JJ. Van de Graaf MW, et al. J Physiol. 2024 Sep;602(18):4529-4542. doi: 10.1113/JP284616. Epub 2023 May 30. J Physiol. 2024. PMID: 37199469 - Data- and knowledge-based modeling of gene regulatory networks: an update.
Linde J, Schulze S, Henkel SG, Guthke R. Linde J, et al. EXCLI J. 2015 Mar 2;14:346-78. doi: 10.17179/excli2015-168. eCollection 2015. EXCLI J. 2015. PMID: 27047314 Free PMC article. Review.
References
- Bar-Joseph Z, et al. Studying and modelling dynamic biological processes using time-series gene expression data. Nat. Rev. Genet. 2012;13:552–564. - PubMed
- Bonneau R, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–1365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- PN2 EY016586/EY/NEI NIH HHS/United States
- U54 CA143907/CA/NCI NIH HHS/United States
- RC1 AI087266/AI/NIAID NIH HHS/United States
- RC4 AI092765/AI/NIAID NIH HHS/United States
- EY016586-06/EY/NEI NIH HHS/United States
- PN1 EY016586/EY/NEI NIH HHS/United States
- IU54CA143907-01/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous