Structure of the tubulin/FtsZ-like protein TubZ from Pseudomonas bacteriophage ΦKZ - PubMed (original) (raw)
Structure of the tubulin/FtsZ-like protein TubZ from Pseudomonas bacteriophage ΦKZ
Christopher H S Aylett et al. J Mol Biol. 2013.
Abstract
Pseudomonas ΦKZ-like bacteriophages encode a group of related tubulin/FtsZ-like proteins believed to be essential for the correct centring of replicated bacteriophage virions within the bacterial host. In this study, we present crystal structures of the tubulin/FtsZ-like protein TubZ from Pseudomonas bacteriophage ΦKZ in both the monomeric and protofilament states, revealing that ΦKZ TubZ undergoes structural changes required to polymerise, forming a canonical tubulin/FtsZ-like protofilament. Combining our structures with previous work, we propose a polymerisation-depolymerisation cycle for the Pseudomonas bacteriophage subgroup of tubulin/FtsZ-like proteins. Electron cryo-microscopy of ΦKZ TubZ filaments polymerised in vitro implies a long-pitch helical arrangement for the constituent protofilaments. Intriguingly, this feature is shared by the other known subgroup of bacteriophage tubulin/FtsZ-like proteins from Clostridium species, which are thought to be involved in partitioning the genomes of bacteriophages adopting a pseudo-lysogenic life cycle.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
Graphical abstract
Fig. S1
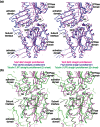
ΦKZ TubZ protofilaments compared to those of Tubulin and FtsZ. (a) Structural superimposition of the protofilament crystal structures of ΦKZ TubZ and FtsZ (PDB ID:
4DXD
). (b) Structural superimposition of the protofilament crystal structures of ΦKZ TubZ and tubulin (PDB ID:
1JFF
). Colour scheme: magenta, polymeric ΦKZ TubZ; blue, FtsZ protofilament; green, tubulin protofilament. All structures are shown as stereographic representations using Cα ribbons.
Fig. 1
Monomeric and protofilament crystal structures of ΦKZ TubZ. (a) Cartoon representation of three subunits of the ΦKZ TubZ protofilament crystal structure; bound GDP shown as spheres. (b) Coomassie-stained SDS-PAGE of ΦKZ TubZ protein; molecular weight standards are expressed in kilodaltons. (c) Cartoon representation of the crystal structure of a monomer of ΦKZ TubZ annotated with the named tubulin/FtsZ secondary structural elements; bound GDP shown as spheres. (d) Cartoon representation of three subunits of the ΦKZ TubZ protofilament crystal structure; bound GDP shown as spheres, rotated 180° and with the central subunit coloured grey in order to highlight the C-terminal knuckle binding site. (e) Structural alignment of the three resolved bacteriophage tubulin/FtsZs. Colour scheme for all plates: green, GTPase domain; yellow, helix 7; magenta, activation domain; cyan, C-terminal helix; nucleotide in cyan and CPK colours.
Fig. 2
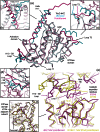
Conformational changes in ΦKZ TubZ during polymerisation. (a) Structural superimposition of the monomeric and protofilament crystal structures of ΦKZ TubZ. (b) Expansion showing the conformational changes undergone by helix 11 and the C-terminus on filament formation. (c) Expansion showing the conformational change undergone by loop T3 on filament formation. (d) Expansion showing the conformational changes undergone by the H10–S9 loop on filament formation. (e) Structural superimposition of the protofilaments of ΦKZ and 201Φ2-1 TubZ (PhuZ) comparing the subunit–subunit interface. (f) Structural superimposition of the C-terminal knuckle regions of ΦKZ and 201Φ2-1 TubZ (PhuZ). Colour scheme: cyan, monomeric ΦKZ TubZ; magenta/purple, ΦKZ TubZ protofilament; yellow, 201Φ2-1 TubZ (PhuZ) protofilament; coloured arrows denote the same region in different structures. All structures are Cα ribbons; distances are expressed in angströms.
Fig. 3
ΦKZ TubZ forms dynamic polymers from intertwined protofilaments. (a) Light scattering of ΦKZ TubZ on addition of GTP. Cyan trace indicates dynamic polymerisation and depolymerisation on addition of 20 μM GTP; magenta trace indicates polymerisation to a plateau on the addition of a saturating concentration of GTP (200 μM). (b) Electron cryo-microscopy of ΦKZ TubZ filaments with saturating concentrations of GTP, showing both polymerised bundles and separated filaments. (c) Fourier transform of a single thick ΦKZ TubZ filament; gyre and pitch layer lines are indicated alongside. (d) Electron micrograph of four single thick ΦKZ TubZ filaments, aligned to highlight the repeat and twist, adjacent to a filtered thick filament produced from the marked layer lines on its Fourier transform.
Similar articles
- Tubulin-Like Proteins in Prokaryotic DNA Positioning.
Fink G, Aylett CHS. Fink G, et al. Subcell Biochem. 2017;84:323-356. doi: 10.1007/978-3-319-53047-5_11. Subcell Biochem. 2017. PMID: 28500531 Review. - Cryo-EM study of the Pseudomonas bacteriophage phiKZ.
Fokine A, Battisti AJ, Bowman VD, Efimov AV, Kurochkina LP, Chipman PR, Mesyanzhinov VV, Rossmann MG. Fokine A, et al. Structure. 2007 Sep;15(9):1099-104. doi: 10.1016/j.str.2007.07.008. Structure. 2007. PMID: 17850749 - Structure of the Bacteriophage PhiKZ Non-virion RNA Polymerase Transcribing from its Promoter p119L.
de Martín Garrido N, Chen CS, Ramlaul K, Aylett CHS, Yakunina M. de Martín Garrido N, et al. J Mol Biol. 2024 Sep 15;436(18):168713. doi: 10.1016/j.jmb.2024.168713. Epub 2024 Jul 17. J Mol Biol. 2024. PMID: 39029888 - Myoviridae bacteriophages of Pseudomonas aeruginosa: a long and complex evolutionary pathway.
Krylov V, Pleteneva E, Bourkaltseva M, Shaburova O, Volckaert G, Sykilinda N, Kurochkina L, Mesyanzhinov V. Krylov V, et al. Res Microbiol. 2003 May;154(4):269-75. doi: 10.1016/S0923-2508(03)00070-6. Res Microbiol. 2003. PMID: 12798231 - Tubulin and FtsZ form a distinct family of GTPases.
Nogales E, Downing KH, Amos LA, Löwe J. Nogales E, et al. Nat Struct Biol. 1998 Jun;5(6):451-8. doi: 10.1038/nsb0698-451. Nat Struct Biol. 1998. PMID: 9628483 Review.
Cited by
- Inside out: tubulin cytomotive filaments versus microtubules.
Sui H. Sui H. Structure. 2014 Apr 8;22(4):509-10. doi: 10.1016/j.str.2014.03.007. Structure. 2014. PMID: 24717557 Free PMC article. - Intracellular Organization by Jumbo Bacteriophages.
Guan J, Bondy-Denomy J. Guan J, et al. J Bacteriol. 2020 Dec 18;203(2):e00362-20. doi: 10.1128/JB.00362-20. Print 2020 Dec 18. J Bacteriol. 2020. PMID: 32868402 Free PMC article. Review. - Bacterial tubulin TubZ-Bt transitions between a two-stranded intermediate and a four-stranded filament upon GTP hydrolysis.
Montabana EA, Agard DA. Montabana EA, et al. Proc Natl Acad Sci U S A. 2014 Mar 4;111(9):3407-12. doi: 10.1073/pnas.1318339111. Epub 2014 Feb 18. Proc Natl Acad Sci U S A. 2014. PMID: 24550513 Free PMC article. - Identifying the core genome of the nucleus-forming bacteriophage family and characterization of Erwinia phage RAY.
Prichard A, Lee J, Laughlin TG, Lee A, Thomas KP, Sy AE, Spencer T, Asavavimol A, Cafferata A, Cameron M, Chiu N, Davydov D, Desai I, Diaz G, Guereca M, Hearst K, Huang L, Jacobs E, Johnson A, Kahn S, Koch R, Martinez A, Norquist M, Pau T, Prasad G, Saam K, Sandhu M, Sarabia AJ, Schumaker S, Sonin A, Uyeno A, Zhao A, Corbett KD, Pogliano K, Meyer J, Grose JH, Villa E, Dutton R, Pogliano J. Prichard A, et al. Cell Rep. 2023 May 30;42(5):112432. doi: 10.1016/j.celrep.2023.112432. Epub 2023 Apr 28. Cell Rep. 2023. PMID: 37120812 Free PMC article. - Bacteriophage genome engineering with CRISPR-Cas13a.
Guan J, Oromí-Bosch A, Mendoza SD, Karambelkar S, Berry JD, Bondy-Denomy J. Guan J, et al. Nat Microbiol. 2022 Dec;7(12):1956-1966. doi: 10.1038/s41564-022-01243-4. Epub 2022 Oct 31. Nat Microbiol. 2022. PMID: 36316452 Free PMC article.
References
- Bergh O., Børsheim K.Y., Bratbak G., Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. - PubMed
- Löwe J., Amos L.A. Evolution of cytomotive filaments: the cytoskeleton from prokaryotes to eukaryotes. Int. J. Biochem. Cell Biol. 2009;41:323–329. - PubMed
- Aylett C.H.S., Löwe J., Amos L.A. New insights into the mechanisms of cytomotive actin and tubulin filaments. Int. Rev. Cell Mol. Biol. 2011;292:1–71. - PubMed
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J. Mol. Biol. 1983;169:353–372. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_U105184313/MRC_/Medical Research Council/United Kingdom
- MC_U105184326/MRC_/Medical Research Council/United Kingdom
- U105184326/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources