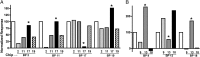Viral infection modulation and neutralization by camelid nanobodies - PubMed (original) (raw)
Viral infection modulation and neutralization by camelid nanobodies
Aline Desmyter et al. Proc Natl Acad Sci U S A. 2013.
Abstract
Lactococcal phages belong to a large family of Siphoviridae and infect Lactococcus lactis, a gram-positive bacterium used in commercial dairy fermentations. These phages are believed to recognize and bind specifically to pellicle polysaccharides covering the entire bacterium. The phage TP901-1 baseplate, located at the tip of the tail, harbors 18 trimeric receptor binding proteins (RBPs) promoting adhesion to a specific lactococcal strain. Phage TP901-1 adhesion does not require major conformational changes or Ca(2+), which contrasts other lactococcal phages. Here, we produced and characterized llama nanobodies raised against the purified baseplate and the Tal protein of phage TP901-1 as tools to dissect the molecular determinants of phage TP901-1 infection. Using a set of complementary techniques, surface plasmon resonance, EM, and X-ray crystallography in a hybrid approach, we identified binders to the three components of the baseplate, analyzed their affinity for their targets, and determined their epitopes as well as their functional impact on TP901-1 phage infectivity. We determined the X-ray structures of three nanobodies in complex with the RBP. Two of them bind to the saccharide binding site of the RBP and are able to fully neutralize TP901-1 phage infectivity, even after 15 passages. These results provide clear evidence for a practical use of nanobodies in circumventing lactococcal phages viral infection in dairy fermentation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Phage TP901-1, its baseplate, and the baseplate components used in this study. (A) The complete phage TP901-1 (8) and the baseplate (brown) at the tip of the tail. (B) The baseplate and the long tripod. (C) The BppL/RBP trimer. (D) The short tripod (three BppU-Ct + nine BppL/RBP).
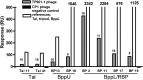
Fig. 2.
SPR analysis of the interactions between nanobodies and TP901-1 virion or phage components. Binding (expressed in response units) of phage TP901-1 virion to the immobilized nanobodies (gray). The positive controls (white) are performed with Tal-Nt, the long tripod, and Bpp for the nanobodies against Tal-Nt, BppU, or BppL as indicated. The negative controls (black) are performed with phage Cp-1.
Fig. 3.
Epitope mapping of the anti-BPTP901-1 nanobodies performed by SPR. (A) Anti-BppL nanobodies. (B) Anti-BppU nanobodies. (A) BppL and (B) BppU were injected over the immobilized nanobodies (Chip) as positive controls (white) and have been normalized to 100. Then, (A) BppL and (B) BppU were injected in presence of the different anti-BppL and anti-BppU nanobodies, and the response signal was normalized to the positive control. *Nanobodies presenting a response pattern dissimilar to the other patterns.
Fig. 4.
Specificity of nanobodies to expressed phage components of TP901-1 vs. Tuc2009 analyzed by SPR. TP901-1 tripod and BppL were injected as a control (white) over the immobilized anti-BppU (Left) and anti-BppL (Right) nanobodies, and the response signal was normalized to 100. Tuc2009 tripod was injected over the immobilized nanobodies (gray), and the response was normalized to the positive control (black letters). For clarity, the response of Tuc2009 tripod vs. anti-BppL nanobodies is rationalized to the molecular weight (gray letters).
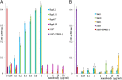
Fig. 5.
Negative staining single-particle EM analysis of the binding of the various nanobodies to the BPTP901-1. (A) The ribbon view of the BPTP901-1 X-ray structure (6) and the difference maps calculated for nanobodies 2, 11, 17, and 19 (anti-BppL) and nanobodies 9, 13, and 18 (anti-BppU). (B) Section observed at the level of the BppU/BppL junction. (C) Section observed at the level of the BppL head domain. (D) Section observed at the level of the BppL stem domain. (E) Close-up view of the BppU/BppL junction. (F) Close-up view of the BppL area.
Fig. 6.
X-ray structures of the various complexes formed by BppL and nanobodies 2, 11, and 17. (A) Ribbon view of the superposition of the three structures of the complexes; (B) 90° view (compared with A) of the BppL complexes with nanobodies 2 and 11 sliced at the level of the head domains. (C) Detailed view of the interaction area between BppL head domains (pink and green surfaces) and CDR2 and -3 of nanobodies 2 (white sticks) and 11 (yellow sticks); (D) 90° view (compared with A) of the BppL complexes with nanobody 17 sliced at the level of the stem domains. (E) Detailed view of the interaction area between BppL stem and β-helix domains (beige surface) and CDR3 of nanobody 17 (green sticks). In C and E, numbering is according to the work by Kabat et al. (47)
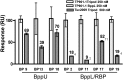
Fig. 7.
Neutralization study of L. lactis strain 3107 infection by phage TP901-1 in the presence of nanobodies. (A) OD at 500 nm of the culture in the presence of 105 pfu TP9010-1 and BppL VHH2, -11, -17, and -19 across the concentration range of 0.05–50 µg/mL after 7 h coincubation at 30 °C. (B) The same conditions were applied with Tal VHH11, -18, and -41. Controls of uninfected and infected culture in the absence of nanobodies were included, and all results are the average of at least triplicate assays.
Similar articles
- The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Legrand P, Collins B, Blangy S, Murphy J, Spinelli S, Gutierrez C, Richet N, Kellenberger C, Desmyter A, Mahony J, van Sinderen D, Cambillau C. Legrand P, et al. mBio. 2016 Jan 26;7(1):e01781-15. doi: 10.1128/mBio.01781-15. mBio. 2016. PMID: 26814179 Free PMC article. - Structure and molecular assignment of lactococcal phage TP901-1 baseplate.
Bebeacua C, Bron P, Lai L, Vegge CS, Brøndsted L, Spinelli S, Campanacci V, Veesler D, van Heel M, Cambillau C. Bebeacua C, et al. J Biol Chem. 2010 Dec 10;285(50):39079-86. doi: 10.1074/jbc.M110.175646. Epub 2010 Oct 11. J Biol Chem. 2010. PMID: 20937834 Free PMC article. - Structural Insights into Lactococcal Siphophage p2 Baseplate Activation Mechanism.
Spinelli S, Tremblay D, Moineau S, Cambillau C, Goulet A. Spinelli S, et al. Viruses. 2020 Aug 11;12(8):878. doi: 10.3390/v12080878. Viruses. 2020. PMID: 32796652 Free PMC article. - Molecular Basis of Bacterial Host Interactions by Gram-Positive Targeting Bacteriophages.
Dunne M, Hupfeld M, Klumpp J, Loessner MJ. Dunne M, et al. Viruses. 2018 Jul 28;10(8):397. doi: 10.3390/v10080397. Viruses. 2018. PMID: 30060549 Free PMC article. Review. - Conserved and Diverse Traits of Adhesion Devices from Siphoviridae Recognizing Proteinaceous or Saccharidic Receptors.
Goulet A, Spinelli S, Mahony J, Cambillau C. Goulet A, et al. Viruses. 2020 May 6;12(5):512. doi: 10.3390/v12050512. Viruses. 2020. PMID: 32384698 Free PMC article. Review.
Cited by
- Potential therapeutic targets and promising drugs for combating SARS-CoV-2.
Zhou H, Fang Y, Xu T, Ni WJ, Shen AZ, Meng XM. Zhou H, et al. Br J Pharmacol. 2020 Jul;177(14):3147-3161. doi: 10.1111/bph.15092. Epub 2020 Jun 5. Br J Pharmacol. 2020. PMID: 32368792 Free PMC article. Review. - First insights into the structural features of Ebola virus methyltransferase activities.
Valle C, Martin B, Ferron F, Roig-Zamboni V, Desmyter A, Debart F, Vasseur JJ, Canard B, Coutard B, Decroly E. Valle C, et al. Nucleic Acids Res. 2021 Feb 22;49(3):1737-1748. doi: 10.1093/nar/gkaa1276. Nucleic Acids Res. 2021. PMID: 33503246 Free PMC article. - Structural flexibility of Toscana virus nucleoprotein in the presence of a single-chain camelid antibody.
Papageorgiou N, Baklouti A, Lichière J, Desmyter A, Canard B, Coutard B, Ferron F. Papageorgiou N, et al. Acta Crystallogr D Struct Biol. 2024 Feb 1;80(Pt 2):113-122. doi: 10.1107/S2059798324000196. Epub 2024 Jan 24. Acta Crystallogr D Struct Biol. 2024. PMID: 38265877 Free PMC article. - Production, crystallization and X-ray diffraction analysis of a complex between a fragment of the TssM T6SS protein and a camelid nanobody.
Nguyen VS, Spinelli S, Desmyter A, Le TT, Kellenberger C, Cascales E, Cambillau C, Roussel A. Nguyen VS, et al. Acta Crystallogr F Struct Biol Commun. 2015 Mar;71(Pt 3):266-71. doi: 10.1107/S2053230X15000709. Epub 2015 Feb 19. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 25760699 Free PMC article. - Interactions of the cell-wall glycopolymers of lactic acid bacteria with their bacteriophages.
Chapot-Chartier MP. Chapot-Chartier MP. Front Microbiol. 2014 May 22;5:236. doi: 10.3389/fmicb.2014.00236. eCollection 2014. Front Microbiol. 2014. PMID: 24904550 Free PMC article. Review.
References
- Kostyuchenko VA, et al. Three-dimensional structure of bacteriophage T4 baseplate. Nat Struct Biol. 2003;10(9):688–693. - PubMed
- Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–1795. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous