Influence of metabolism on epigenetics and disease - PubMed (original) (raw)
Review
Influence of metabolism on epigenetics and disease
William G Kaelin Jr et al. Cell. 2013.
Abstract
Chemical modifications of histones and DNA, such as histone methylation, histone acetylation, and DNA methylation, play critical roles in epigenetic gene regulation. Many of the enzymes that add or remove such chemical modifications are known, or might be suspected, to be sensitive to changes in intracellular metabolism. This knowledge provides a conceptual foundation for understanding how mutations in the metabolic enzymes SDH, FH, and IDH can result in cancer and, more broadly, for how alterations in metabolism and nutrition might contribute to disease. Here, we review literature pertinent to hypothetical connections between metabolic and epigenetic states in eukaryotic cells.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
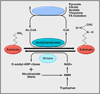
Figure 1. Metabolism and Acetylation/Deacetylation
Histone acetylases use acetyl-CoA (Ac-CoA) as an acetyl donor, whose synthesis requires coenzyme A (CoA). Ac-CoA canbe regeneratedin chemical reactions involving pyruvate, citrate, acetate, and various amino acids such as threonine and by fatty acid beta oxidation. Deacetylation by Sirtuin family histone deacetylases requires NAD+, leading to the generation of O-acetyl-ADP ribose and nicotinamide (NAM). NAD+ is produced from NMN (nicotinamide mononucleotide), which can be salvaged from NAM or produced de novo from tryptophan. For simplicity, enzymes catalyzing the various reactions are not shown.
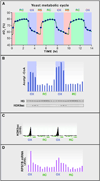
Figure 2. Evidence of Transient Acetylation of Histone H3 Only during the Oxidative Phase of the Yeast Metabolic Cycle
(A) Periodic fluctuation in oxygen levels in a chemostat growing prototrophic yeast. The yeast metabolic cycle (YMC) is roughly 5 hours in duration and defined by sequentially repeating oxidative (Ox), reductive building (RB), and reductive charging (RC) metabolic phases (adapted from Tu et al., 2005). (B) Quantitative measurement of acetyl-CoA levels over the YMC reveal elevated levels of the metabolite during the Ox phase of the YMC. Western blot measurements of H3K9 acetylation over the YMC reveal dynamic acetylation temporally correlate with the peak abundance of acetyl-CoA. (C) ChIP-seq analysis of H3K9 acetylation on the promoter of the gene encoding the RPS7B ribosomal protein reveals modification limited to the Ox phase of the YMC. (D) Transcript abundance of the RPS7B mRNA peaks during the Ox phase of the YMC precisely when acetyl-CoA levels are of highest abundance and when the promoter of the RPS7B gene is modified by H3K9 acetylation.
Figure 3. Metabolism and Methyltrans-ferases
DNA and histone methyltransferases use S-adenosylmethionine (SAM), derived from methionine, as a methyl donor, resulting in the generation of S-adenosylhomocysteine (SAH). SAH is converted to homocysteine, which is then converted back to methionine in a vitamin B12-dependent reaction that utilizes carbons derived from either choline or folate. DHF, dihydrofolate; THF, tetrahydrofolate; 5,10-MTHF, 5,10-methylene THF; CH3, methyl. Also shown are steps requiring vitamin B6 and B2. For simplicity, enzymes catalyzing the various reactions are not shown.
Figure 4. TDH-Mediated Catabolism of Threonine
Threonine is catabolized to acetyl-CoA and glycine via a two-step process, first involving the rate-limiting threonine dehydrogenase (TDH) enzyme yielding the short-lived intermediate 2-amino-3-ketobutyrate. This intermediate is subsequently subject to the 2-amino-3-ketobutyrate ligase (KBL) enzyme that, supplemented by coenzyme A (CoA), yields the final products of the reaction, acetyl-CoA and glycine. Both steps of the catabolic reaction take place in the mitochondria of eukaryotic cells. The former product, acetyl-CoA,can be fed into the TCA cycle or used as an anabolic building block for other metabolites. The latter product, glycine, is used to feed the mitochondrial glycine cleavage system for the conversion of tetrahydrofolate (THF) into N5, N10-methylene-tetrahydrofolate (MTHF). MTHF, in turn, is capable of one-carbon donation in biosynthetic reactions involving purine and pyrimidine synthesis, as well as the regeneration of methionine from homocysteine.
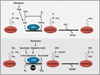
Figure 5. Histone Demethylase Reactions
LSD demethylases oxidize monomethylated and dimethylated histones using FAD (flavin adenine dinucleotide) as a cofactor. The oxidized methyl group is unstable and, after attack by water, given off as formaldehyde (HCHO). FAD is derived from FMN (flavin mononucleotide), which in turn is derived from riboflavin. JmjC demethylases hydroxylate methylated histones in a reaction coupled to decarboxylation of 2-oxoglutarate to succinate. 2-oxoglutarate (also called α-ketoglutarate) can be derived from several sources including isocitrate and glutamic acid. Spontaneous release of the hydroxylated methyl group results in demethylation.
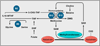
Figure 6. Mutant Metabolic Enzymes, Epigenetics, and Cancer
(A) The abundance of the HIF transcription factor is regulated by the 2-oxoglutarate-dependent EglN dioxygenases, which are sensitive to changes in oxygen and metabolism, and by the PI3K–AKT-mTOR pathway, which is involved in nutrient sensing and frequently mutationally activated in cancer. HIF induces the transcription of a number of JmjC histone demethylases. (B) Mutational inactivation of the Krebs Cycle Enzymes SDH and FH leads to the accumulation of succinate and fumarate, respectively, whereas tumor-derived IDH1 and IDH2 mutants produce high levels of R-2-hydroxyglutarate (R-2HG). Succinate, fumarate, and R-2HG can inhibit 2-oxoglutarate-dependent dioxy-genases, including JmjC histone demethylases and TET DNA hydroxylases.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - Dynamic Field Theory of Executive Function: Identifying Early Neurocognitive Markers.
McCraw A, Sullivan J, Lowery K, Eddings R, Heim HR, Buss AT. McCraw A, et al. Monogr Soc Res Child Dev. 2024 Dec;89(3):7-109. doi: 10.1111/mono.12478. Monogr Soc Res Child Dev. 2024. PMID: 39628288 Free PMC article. - Australia in 2030: what is our path to health for all?
Backholer K, Baum F, Finlay SM, Friel S, Giles-Corti B, Jones A, Patrick R, Shill J, Townsend B, Armstrong F, Baker P, Bowen K, Browne J, Büsst C, Butt A, Canuto K, Canuto K, Capon A, Corben K, Daube M, Goldfeld S, Grenfell R, Gunn L, Harris P, Horton K, Keane L, Lacy-Nichols J, Lo SN, Lovett RW, Lowe M, Martin JE, Neal N, Peeters A, Pettman T, Thoms A, Thow AMT, Timperio A, Williams C, Wright A, Zapata-Diomedi B, Demaio S. Backholer K, et al. Med J Aust. 2021 May;214 Suppl 8:S5-S40. doi: 10.5694/mja2.51020. Med J Aust. 2021. PMID: 33934362 - A systematic review of epigenetic interplay in kidney diseases: Crosstalk between long noncoding RNAs and methylation, acetylation of chromatin and histone.
Tan RZ, Jia J, Li T, Wang L, Kantawong F. Tan RZ, et al. Biomed Pharmacother. 2024 Jul;176:116922. doi: 10.1016/j.biopha.2024.116922. Epub 2024 Jun 12. Biomed Pharmacother. 2024. PMID: 38870627 Review.
Cited by
- Bidirectional interplay between metabolism and epigenetics in hematopoietic stem cells and leukemia.
Zhang YW, Schönberger K, Cabezas-Wallscheid N. Zhang YW, et al. EMBO J. 2023 Dec 11;42(24):e112348. doi: 10.15252/embj.2022112348. Epub 2023 Nov 27. EMBO J. 2023. PMID: 38010205 Free PMC article. Review. - The metabolic role of the CD73/adenosine signaling pathway in HTR-8/SVneo cells: A Double-Edged Sword?
Song G, Zhang D, Zhu J, Wang A, Zhou X, Han TL, Zhang H. Song G, et al. Heliyon. 2024 Jan 29;10(3):e25252. doi: 10.1016/j.heliyon.2024.e25252. eCollection 2024 Feb 15. Heliyon. 2024. PMID: 38322906 Free PMC article. - The regulation of HBP1, SIRT1, and SREBP-1c genes and the related microRNAs in non-alcoholic fatty liver rats: The association with the folic acid anti-steatosis.
Salman M, Kamel MA, El-Nabi SEH, Ismail AHA, Ullah S, Al-Ghamdi A, Hathout HMR, El-Garawani IM. Salman M, et al. PLoS One. 2022 Apr 13;17(4):e0265455. doi: 10.1371/journal.pone.0265455. eCollection 2022. PLoS One. 2022. PMID: 35417465 Free PMC article. - Sensitization of Resistant Breast Cancer Cells with a Jumonji Family Histone Demethylase Inhibitor.
Singh B, Sarli VN, Lucci A. Singh B, et al. Cancers (Basel). 2022 May 26;14(11):2631. doi: 10.3390/cancers14112631. Cancers (Basel). 2022. PMID: 35681611 Free PMC article. - Loss of EZH2 Reprograms BCAA Metabolism to Drive Leukemic Transformation.
Gu Z, Liu Y, Cai F, Patrick M, Zmajkovic J, Cao H, Zhang Y, Tasdogan A, Chen M, Qi L, Liu X, Li K, Lyu J, Dickerson KE, Chen W, Ni M, Merritt ME, Morrison SJ, Skoda RC, DeBerardinis RJ, Xu J. Gu Z, et al. Cancer Discov. 2019 Sep;9(9):1228-1247. doi: 10.1158/2159-8290.CD-19-0152. Epub 2019 Jun 12. Cancer Discov. 2019. PMID: 31189531 Free PMC article.
References
- Adam J, Hatipoglu E, O’Flaherty L, Ternette N, Sahgal N, Lockstone H, Baban D, Nye E, Stamp GW, Wolhuter K, et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell. 2011;20:524–537. - PMC - PubMed
- Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat. Genet. 2011;43:1262–1265. - PubMed
- Andreeva VA, Touvier M, Kesse-Guyot E, Julia C, Galan P, Hercberg S. B vitamin and/or u-3 fatty acid supplementation and cancer: ancillary findings from the supplementation with folate, vitamins B6 and B12, and/or omega-3 fatty acids (SU.FOL.OM3) randomized trial. Arch. Intern. Med. 2012;172:540–547. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous