Coalescent-based genome analyses resolve the early branches of the euarchontoglires - PubMed (original) (raw)
Coalescent-based genome analyses resolve the early branches of the euarchontoglires
Vikas Kumar et al. PLoS One. 2013.
Abstract
Despite numerous large-scale phylogenomic studies, certain parts of the mammalian tree are extraordinarily difficult to resolve. We used the coding regions from 19 completely sequenced genomes to study the relationships within the super-clade Euarchontoglires (Primates, Rodentia, Lagomorpha, Dermoptera and Scandentia) because the placement of Scandentia within this clade is controversial. The difficulty in resolving this issue is due to the short time spans between the early divergences of Euarchontoglires, which may cause incongruent gene trees. The conflict in the data can be depicted by network analyses and the contentious relationships are best reconstructed by coalescent-based analyses. This method is expected to be superior to analyses of concatenated data in reconstructing a species tree from numerous gene trees. The total concatenated dataset used to study the relationships in this group comprises 5,875 protein-coding genes (9,799,170 nucleotides) from all orders except Dermoptera (flying lemurs). Reconstruction of the species tree from 1,006 gene trees using coalescent models placed Scandentia as sister group to the primates, which is in agreement with maximum likelihood analyses of concatenated nucleotide sequence data. Additionally, both analytical approaches favoured the Tarsier to be sister taxon to Anthropoidea, thus belonging to the Haplorrhine clade. When divergence times are short such as in radiations over periods of a few million years, even genome scale analyses struggle to resolve phylogenetic relationships. On these short branches processes such as incomplete lineage sorting and possibly hybridization occur and make it preferable to base phylogenomic analyses on coalescent methods.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
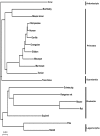
Figure 1. The ML tree of concatenated nucleotides data from 5,875 genes with all the branches being unanimously supported by TF.
Figure 2. The ML tree based on amino acid data from 5,875 genes representing the best option for a bifurcating topology.
Only the TF support values <99 are shown.
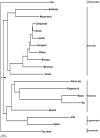
Figure 3. Species tree based on 1,006 gene trees with bootstrap support values (>99% not shown).
Above value indicates the STAR support value and MP-EST values are indicated below.
Figure 4. Consensus network in which at least 10% of the 1006 ML gene trees have common branches (threshold value 10%).
Similar articles
- The gene tree delusion.
Springer MS, Gatesy J. Springer MS, et al. Mol Phylogenet Evol. 2016 Jan;94(Pt A):1-33. doi: 10.1016/j.ympev.2015.07.018. Epub 2015 Jul 31. Mol Phylogenet Evol. 2016. PMID: 26238460 - Flying lemurs--the 'flying tree shrews'? Molecular cytogenetic evidence for a Scandentia-Dermoptera sister clade.
Nie W, Fu B, O'Brien PC, Wang J, Su W, Tanomtong A, Volobouev V, Ferguson-Smith MA, Yang F. Nie W, et al. BMC Biol. 2008 May 1;6:18. doi: 10.1186/1741-7007-6-18. BMC Biol. 2008. PMID: 18452598 Free PMC article. - Phylogenetic analyses of complete mitochondrial genome sequences suggest a basal divergence of the enigmatic rodent Anomalurus.
Horner DS, Lefkimmiatis K, Reyes A, Gissi C, Saccone C, Pesole G. Horner DS, et al. BMC Evol Biol. 2007 Feb 8;7:16. doi: 10.1186/1471-2148-7-16. BMC Evol Biol. 2007. PMID: 17288612 Free PMC article. - Molecular cytogenetic studies in strepsirrhine primates, Dermoptera and Scandentia.
Nie W. Nie W. Cytogenet Genome Res. 2012;137(2-4):246-58. doi: 10.1159/000338727. Epub 2012 May 17. Cytogenet Genome Res. 2012. PMID: 22614467 Review. - [Molecular evidence on the phylogenetic position of tree shrews].
Xu L, Fan Y, Jiang XL, Yao YG. Xu L, et al. Dongwuxue Yanjiu. 2013 Apr;34(2):70-6. doi: 10.3724/SP.J.1141.2013.02070. Dongwuxue Yanjiu. 2013. PMID: 23572355 Review. Chinese.
Cited by
- Phylogenomic analyses of nuclear genes reveal the evolutionary relationships within the BEP clade and the evidence of positive selection in Poaceae.
Zhao L, Zhang N, Ma PF, Liu Q, Li DZ, Guo ZH. Zhao L, et al. PLoS One. 2013 May 29;8(5):e64642. doi: 10.1371/journal.pone.0064642. Print 2013. PLoS One. 2013. PMID: 23734211 Free PMC article. - Global abundance of short tandem repeats is non-random in rodents and primates.
Arabfard M, Salesi M, Nourian YH, Arabipour I, Maddi AA, Kavousi K, Ohadi M. Arabfard M, et al. BMC Genom Data. 2022 Nov 3;23(1):77. doi: 10.1186/s12863-022-01092-4. BMC Genom Data. 2022. PMID: 36329409 Free PMC article. - Comparison of the Microsatellite Distribution Patterns in the Genomes of Euarchontoglires at the Taxonomic Level.
Song X, Yang T, Zhang X, Yuan Y, Yan X, Wei Y, Zhang J, Zhou C. Song X, et al. Front Genet. 2021 Feb 26;12:622724. doi: 10.3389/fgene.2021.622724. eCollection 2021. Front Genet. 2021. PMID: 33719337 Free PMC article. - Disentangling genetic structure for genetic monitoring of complex populations.
Milligan BG, Archer FI, Ferchaud AL, Hand BK, Kierepka EM, Waples RS. Milligan BG, et al. Evol Appl. 2018 Mar 23;11(7):1149-1161. doi: 10.1111/eva.12622. eCollection 2018 Aug. Evol Appl. 2018. PMID: 30026803 Free PMC article. - Conserved Signatures in Protein Sequences Reliably Demarcate Different Clades of Rodents/Glires Species and Consolidate Their Evolutionary Relationships.
Gupta RS, Suggett C. Gupta RS, et al. Genes (Basel). 2022 Feb 1;13(2):288. doi: 10.3390/genes13020288. Genes (Basel). 2022. PMID: 35205335 Free PMC article.
References
- Hallström BM, Kullberg M, Nilsson MA, Janke A (2007) Phylogenomic data analyses provide evidence that Xenarthra and Afrotheria are sister groups. Mol Biol Evol 24: 2059–2068. - PubMed
Publication types
MeSH terms
Grants and funding
The present study was supported by the research funding program LOEWE: "Landes-Offensive zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz" of Hesse's Ministry of Higher Education, Research, and the Arts. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources