Surprisingly extensive mixed phylogenetic and ecological signals among bacterial Operational Taxonomic Units - PubMed (original) (raw)
. 2013 May 1;41(10):5175-88.
doi: 10.1093/nar/gkt241. Epub 2013 Apr 9.
Affiliations
- PMID: 23571758
- PMCID: PMC3664822
- DOI: 10.1093/nar/gkt241
Surprisingly extensive mixed phylogenetic and ecological signals among bacterial Operational Taxonomic Units
Alexander F Koeppel et al. Nucleic Acids Res. 2013.
Abstract
The lack of a consensus bacterial species concept greatly hampers our ability to understand and organize bacterial diversity. Operational taxonomic units (OTUs), which are clustered on the basis of DNA sequence identity alone, are the most commonly used microbial diversity unit. Although it is understood that OTUs can be phylogenetically incoherent, the degree and the extent of the phylogenetic inconsistency have not been explicitly studied. Here, we tested the phylogenetic signal of OTUs in a broad range of bacterial genera from various phyla. Strikingly, we found that very few OTUs were monophyletic, and many showed evidence of multiple independent origins. Using previously established bacterial habitats as benchmarks, we showed that OTUs frequently spanned multiple ecological habitats. We demonstrated that ecological heterogeneity within OTUs is caused by their phylogenetic inconsistency, and not merely due to 'lumping' of taxa resulting from using relaxed identity cut-offs. We argue that ecotypes, as described by the Stable Ecotype Model, are phylogenetically and ecologically more consistent than OTUs and therefore could serve as an alternative unit for bacterial diversity studies. In addition, we introduce QuickES, a new wrapper program for the Ecotype Simulation algorithm, which is capable of demarcating ecotypes in data sets with tens of thousands of sequences.
Figures
Figure 1.
OTU paraphyly is pervasive and pronounced. This graph plots OTU size against PI for all 99% 16S rRNA OTUs among 10 genera. PI values of 0.0 indicate monophyletic groups, whereas a PI close to 1 indicates substantial paraphyly. Genus classifications of OTUs are colour coded as indicated in the key.
Figure 2.
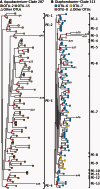
Extensive paraphyly and polyphyly among OTUs. Maximum likelihood trees of representative subclades of the genera (A) Aquabacterium and (B) Diaphorobacter. OTU generated using the 99% identity cut-off are shown with the putative ecotypes (PE) demarcated by ES. Internal nodes with >80% bootstrap support are highlighted with red circles.
Figure 3.
OTUs and Ecotypes show distinct habitat associations. An ML tree of a subset of the Vibrio hsp60 sequences (A) and a neighbour-joining tree of the full set of Synechococcus psaA sequences (B). OTUs, ES ecotypes and AdaptML ecotypes are shown. Note that the formatting in the OTU column and the AdaptML column is different. In the OTU column, all leaves marked by the same color belong to the same OTU. In the AdaptML column, different colours denoted different habitats. Each distinct colour bar is its own ecotype, whereas bars of the same colour are ecotypes co-occurring in the same habitat.
Figure 4.
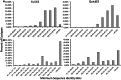
Putative ecotypes are not captured by any single sequence identity cut-off. Graphs display the number of ecotypes whose minimum pairwise sequence identity fall into each of the displayed bins in the skin Aquabacterium (A) and the marine Vibrio (B) data sets.
Similar articles
- Species matter: the role of competition in the assembly of congeneric bacteria.
Koeppel AF, Wu M. Koeppel AF, et al. ISME J. 2014 Mar;8(3):531-540. doi: 10.1038/ismej.2013.180. Epub 2013 Oct 17. ISME J. 2014. PMID: 24132075 Free PMC article. - A meta-analysis of the publicly available bacterial and archaeal sequence diversity in saline soils.
Ma B, Gong J. Ma B, et al. World J Microbiol Biotechnol. 2013 Dec;29(12):2325-34. doi: 10.1007/s11274-013-1399-9. Epub 2013 Jun 12. World J Microbiol Biotechnol. 2013. PMID: 23756871 - The quorum sensing diversity within and between ecotypes of Bacillus subtilis.
Stefanic P, Decorosi F, Viti C, Petito J, Cohan FM, Mandic-Mulec I. Stefanic P, et al. Environ Microbiol. 2012 Jun;14(6):1378-89. doi: 10.1111/j.1462-2920.2012.02717.x. Epub 2012 Mar 6. Environ Microbiol. 2012. PMID: 22390407 - Tracking bacterial responses to global warming with an ecotype-based systematics.
Cohan FM. Cohan FM. Clin Microbiol Infect. 2009 Jan;15 Suppl 1:54-9. doi: 10.1111/j.1469-0691.2008.02681.x. Clin Microbiol Infect. 2009. PMID: 19220357 Review. - Bacterial species and speciation.
Cohan FM. Cohan FM. Syst Biol. 2001 Aug;50(4):513-24. doi: 10.1080/10635150118398. Syst Biol. 2001. PMID: 12116650 Review.
Cited by
- Taxonomy Informed Clustering, an Optimized Method for Purer and More Informative Clusters in Diversity Analysis and Microbiome Profiling.
Kioukis A, Pourjam M, Neuhaus K, Lagkouvardos I. Kioukis A, et al. Front Bioinform. 2022 Apr 27;2:864597. doi: 10.3389/fbinf.2022.864597. eCollection 2022. Front Bioinform. 2022. PMID: 36304326 Free PMC article. - The human gut pan-microbiome presents a compositional core formed by discrete phylogenetic units.
Aguirre de Cárcer D. Aguirre de Cárcer D. Sci Rep. 2018 Sep 19;8(1):14069. doi: 10.1038/s41598-018-32221-8. Sci Rep. 2018. PMID: 30232462 Free PMC article. - Weak Coherence in Abundance Patterns Between Bacterial Classes and Their Constituent OTUs Along a Regulated River.
Ruiz-González C, Salazar G, Logares R, Proia L, Gasol JM, Sabater S. Ruiz-González C, et al. Front Microbiol. 2015 Nov 26;6:1293. doi: 10.3389/fmicb.2015.01293. eCollection 2015. Front Microbiol. 2015. PMID: 26635761 Free PMC article. - Microbial Community Field Surveys Reveal Abundant Pseudomonas Population in Sorghum Rhizosphere Composed of Many Closely Related Phylotypes.
Chiniquy D, Barnes EM, Zhou J, Hartman K, Li X, Sheflin A, Pella A, Marsh E, Prenni J, Deutschbauer AM, Schachtman DP, Tringe SG. Chiniquy D, et al. Front Microbiol. 2021 Mar 9;12:598180. doi: 10.3389/fmicb.2021.598180. eCollection 2021. Front Microbiol. 2021. PMID: 33767674 Free PMC article. - Microdiversity ensures the maintenance of functional microbial communities under changing environmental conditions.
García-García N, Tamames J, Linz AM, Pedrós-Alió C, Puente-Sánchez F. García-García N, et al. ISME J. 2019 Dec;13(12):2969-2983. doi: 10.1038/s41396-019-0487-8. Epub 2019 Aug 16. ISME J. 2019. PMID: 31417155 Free PMC article.
References
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the sargasso sea. Science. 2004;304:66–74. - PubMed
- Giovannoni SJ, Stingl U. Molecular diversity and ecology of microbial plankton. Nature. 2005;437:343–348. - PubMed
- Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources