New players to the field of ADP-ribosylation make the final cut - PubMed (original) (raw)
Comment
New players to the field of ADP-ribosylation make the final cut
Jamin D Steffen et al. EMBO J. 2013.
Abstract
EMBO J (2013) 32 9, 1225–1237. doi:; DOI: 10.1038/emboj.2013.51
ADP-ribose-based intermediates, including PARP-generated mono- and poly(ADP-ribose) post-translational modifications, are important to a number of cellular signalling processes. The reversal of poly(ADP-ribosyl)ation is mostly attributed to PARG, which however cannot remove the final protein-linked mono(ADP-ribose) residue. Three recent studies, one of them in The EMBO Journal, now report that certain macrodomains remove terminal ADP-ribose modifications from acidic residues.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
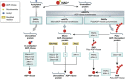
Figure 1
Pathway of reversible protein modification reactions that involve NAD+ consumption. Consumption of NAD+ through nicotinamide cleavage drives the catalysis of mono- and poly(ADP-ribosyl)ation of proteins by ADP-ribosyl transferases, as well as deacetylation of proteins by Sirtuins. Formation of these modifications is important to various cell signalling events such as DNA repair, chromatin remodelling, transcription, telomere homeostasis, and cell death. ADP-ribose modifications are short-lived due to the activity of hydrolase enzymes reversing the modification to yield ADP-ribose. The recently identified macrodomains C6orf130/TARG, MacroD1, and MacroD2 now fill in previously unidentified roles of ADP-ribose and PAR hydrolysis from acidic residues.
Comment on
- Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I. Sharifi R, et al. EMBO J. 2013 May 2;32(9):1225-37. doi: 10.1038/emboj.2013.51. Epub 2013 Mar 12. EMBO J. 2013. PMID: 23481255 Free PMC article.
Similar articles
- Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease.
Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I. Sharifi R, et al. EMBO J. 2013 May 2;32(9):1225-37. doi: 10.1038/emboj.2013.51. Epub 2013 Mar 12. EMBO J. 2013. PMID: 23481255 Free PMC article. - Structures and Mechanisms of Enzymes Employed in the Synthesis and Degradation of PARP-Dependent Protein ADP-Ribosylation.
Barkauskaite E, Jankevicius G, Ahel I. Barkauskaite E, et al. Mol Cell. 2015 Jun 18;58(6):935-46. doi: 10.1016/j.molcel.2015.05.007. Mol Cell. 2015. PMID: 26091342 Review. - [Metabolism and cellular functions of poly(ADP-ribose)].
Miwa M, Masutani M. Miwa M, et al. Seikagaku. 2006 Nov;78(11):1050-61. Seikagaku. 2006. PMID: 17176893 Review. Japanese. No abstract available. - Nucleolar-nucleoplasmic shuttling of TARG1 and its control by DNA damage-induced poly-ADP-ribosylation and by nucleolar transcription.
Bütepage M, Preisinger C, von Kriegsheim A, Scheufen A, Lausberg E, Li J, Kappes F, Feederle R, Ernst S, Eckei L, Krieg S, Müller-Newen G, Rossetti G, Feijs KLH, Verheugd P, Lüscher B. Bütepage M, et al. Sci Rep. 2018 Apr 30;8(1):6748. doi: 10.1038/s41598-018-25137-w. Sci Rep. 2018. PMID: 29712969 Free PMC article. - [Role of poly(ADP-ribosyl)ation in cell death].
Maruta H, Tanuma S. Maruta H, et al. Seikagaku. 2005 Jun;77(6):484-90. Seikagaku. 2005. PMID: 16022423 Review. Japanese. No abstract available.
Cited by
- The rise and fall of poly(ADP-ribose): An enzymatic perspective.
Pascal JM, Ellenberger T. Pascal JM, et al. DNA Repair (Amst). 2015 Aug;32:10-16. doi: 10.1016/j.dnarep.2015.04.008. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25963443 Free PMC article. Review. - Nudix hydrolases degrade protein-conjugated ADP-ribose.
Daniels CM, Thirawatananond P, Ong SE, Gabelli SB, Leung AK. Daniels CM, et al. Sci Rep. 2015 Dec 16;5:18271. doi: 10.1038/srep18271. Sci Rep. 2015. PMID: 26669448 Free PMC article. - An uncharacterized FMAG_01619 protein from Fusobacterium mortiferum ATCC 9817 demonstrates that some bacterial macrodomains can also act as poly-ADP-ribosylhydrolases.
García-Saura AG, Zapata-Pérez R, Hidalgo JF, Cabanes J, Gil-Ortiz F, Sánchez-Ferrer Á. García-Saura AG, et al. Sci Rep. 2019 Mar 1;9(1):3230. doi: 10.1038/s41598-019-39691-4. Sci Rep. 2019. PMID: 30824723 Free PMC article. - Structural and functional analysis of Oceanobacillus iheyensis macrodomain reveals a network of waters involved in substrate binding and catalysis.
Zapata-Pérez R, Gil-Ortiz F, Martínez-Moñino AB, García-Saura AG, Juanhuix J, Sánchez-Ferrer Á. Zapata-Pérez R, et al. Open Biol. 2017 Apr;7(4):160327. doi: 10.1098/rsob.160327. Open Biol. 2017. PMID: 28446708 Free PMC article. - Structural Implications for Selective Targeting of PARPs.
Steffen JD, Brody JR, Armen RS, Pascal JM. Steffen JD, et al. Front Oncol. 2013 Dec 20;3:301. doi: 10.3389/fonc.2013.00301. Front Oncol. 2013. PMID: 24392349 Free PMC article. Review.
References
- Gao H, Coyle DL, Meyer-Ficca ML, Meyer RG, Jacobson EL, Wang ZQ, Jacobson MK (2007) Altered poly(ADP-ribose) metabolism impairs cellular responses to genotoxic stress in a hypomorphic mutant of poly(ADP-ribose) glycohydrolase. Exp Cell Res 313: 984–996 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical