Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians - PubMed (original) (raw)
doi: 10.1371/journal.pone.0059494. Epub 2013 Apr 5.
Jian Li, Jigang Zhang, Chao Xu, Yan Jiang, Zikai Wu, Fuping Zhao, Li Liao, Jun Chen, Yong Lin, Qing Tian, Christopher J Papasian, Hong-Wen Deng
Affiliations
- PMID: 23577066
- PMCID: PMC3618277
- DOI: 10.1371/journal.pone.0059494
Comprehensive characterization of human genome variation by high coverage whole-genome sequencing of forty four Caucasians
Hui Shen et al. PLoS One. 2013.
Abstract
Whole genome sequencing studies are essential to obtain a comprehensive understanding of the vast pattern of human genomic variations. Here we report the results of a high-coverage whole genome sequencing study for 44 unrelated healthy Caucasian adults, each sequenced to over 50-fold coverage (averaging 65.8×). We identified approximately 11 million single nucleotide polymorphisms (SNPs), 2.8 million short insertions and deletions, and over 500,000 block substitutions. We showed that, although previous studies, including the 1000 Genomes Project Phase 1 study, have catalogued the vast majority of common SNPs, many of the low-frequency and rare variants remain undiscovered. For instance, approximately 1.4 million SNPs and 1.3 million short indels that we found were novel to both the dbSNP and the 1000 Genomes Project Phase 1 data sets, and the majority of which (∼96%) have a minor allele frequency less than 5%. On average, each individual genome carried ∼3.3 million SNPs and ∼492,000 indels/block substitutions, including approximately 179 variants that were predicted to cause loss of function of the gene products. Moreover, each individual genome carried an average of 44 such loss-of-function variants in a homozygous state, which would completely "knock out" the corresponding genes. Across all the 44 genomes, a total of 182 genes were "knocked-out" in at least one individual genome, among which 46 genes were "knocked out" in over 30% of our samples, suggesting that a number of genes are commonly "knocked-out" in general populations. Gene ontology analysis suggested that these commonly "knocked-out" genes are enriched in biological process related to antigen processing and immune response. Our results contribute towards a comprehensive characterization of human genomic variation, especially for less-common and rare variants, and provide an invaluable resource for future genetic studies of human variation and diseases.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
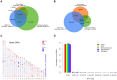
Figure 1. Summary characterizations of the identified variants.
A–B, Venn diagram showing SNPs and indels identified in the present study overlapping with those archived in the dbSNP (v131) and the 1000 Genomes Project Phase 1 data sets (released on 5/21/2011). To account for differences in placement of many indels between different data sets, indels were considered to match if they were within 25 bp distance and of the same size. Only SNPs and indels mapped to autosomes and X chromosome were analyzed. C, Genome-wide distribution of novel SNPs. Total number of novel SNPs (compared to dbSNP v131 and the 1000 Genomes Project pilot phase) were calculated in non-overlap 1-megabases (Mb) windows across the human genome and plotted in ideograms using Idiographica. The diversities were illustrated by colors, with red indicating higher numbers or proportions and blue indicating lower numbers or proportions. Genomic regions in which no SNPs were identified or no reference sequences could be determined are shown in grey. D, Allele frequency spectrum of novel SNPs.
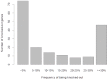
Figure 2. Identification of “knocked-out” genes.
A, Frequency spectrum of observed “knocked-out” genes. Genes containing homozygous LoF variants were expected to be silent or knocked-out. Numbers of “knocked-out” genes were counted with respect to the frequency of “knock-out” occurrence in the 44 genomes.
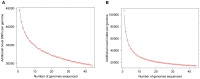
Figure 3. The number of novel SNPs and indels discovered as the number of sequenced genomes increased.
We evaluated how many additional “new” A) SNPs and B) indels, respectively, were identified per genome as the number of sequenced genomes increased, considering both variants archived in databases (dbSNP v131 and the 1000 Genome Project Phase 1 data) and variants “discovered” in previously considered genomes. The 44 genomes were added into the analyses in a random order. With 1000 permutations, the average numbers of novel variants added per genome are shown, along with the best fitting trendline for each plot.
Similar articles
- A map of human genome variation from population-scale sequencing.
1000 Genomes Project Consortium; Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. 1000 Genomes Project Consortium, et al. Nature. 2010 Oct 28;467(7319):1061-73. doi: 10.1038/nature09534. Nature. 2010. PMID: 20981092 Free PMC article. - A global reference for human genetic variation.
1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 1000 Genomes Project Consortium, et al. Nature. 2015 Oct 1;526(7571):68-74. doi: 10.1038/nature15393. Nature. 2015. PMID: 26432245 Free PMC article. - Deep sequencing of Danish Holstein dairy cattle for variant detection and insight into potential loss-of-function variants in protein coding genes.
Das A, Panitz F, Gregersen VR, Bendixen C, Holm LE. Das A, et al. BMC Genomics. 2015 Dec 9;16:1043. doi: 10.1186/s12864-015-2249-y. BMC Genomics. 2015. PMID: 26645365 Free PMC article. - [DNA polymorphisms].
Suehiro Y, Furuya T, Sasaki K, Hinota Y. Suehiro Y, et al. Rinsho Byori. 2013 Nov;61(11):1001-7. Rinsho Byori. 2013. PMID: 24450105 Review. Japanese. - Small insertions and deletions (INDELs) in human genomes.
Mullaney JM, Mills RE, Pittard WS, Devine SE. Mullaney JM, et al. Hum Mol Genet. 2010 Oct 15;19(R2):R131-6. doi: 10.1093/hmg/ddq400. Epub 2010 Sep 21. Hum Mol Genet. 2010. PMID: 20858594 Free PMC article. Review.
Cited by
- Identification of novel functional CpG-SNPs associated with Type 2 diabetes and birth weight.
Liu RK, Lin X, Wang Z, Greenbaum J, Qiu C, Zeng CP, Zhu YY, Shen J, Deng HW. Liu RK, et al. Aging (Albany NY). 2021 Apr 4;13(7):10619-10658. doi: 10.18632/aging.202828. Epub 2021 Apr 4. Aging (Albany NY). 2021. PMID: 33835050 Free PMC article. - Focused Strategies for Defining the Genetic Architecture of Congenital Heart Defects.
Martin LJ, Benson DW. Martin LJ, et al. Genes (Basel). 2021 May 28;12(6):827. doi: 10.3390/genes12060827. Genes (Basel). 2021. PMID: 34071175 Free PMC article. Review. - An Ensemble Approach to Predict the Pathogenicity of Synonymous Variants.
Ranganathan Ganakammal S, Alexov E. Ranganathan Ganakammal S, et al. Genes (Basel). 2020 Sep 21;11(9):1102. doi: 10.3390/genes11091102. Genes (Basel). 2020. PMID: 32967157 Free PMC article. - DNA copy number variation: Main characteristics, evolutionary significance, and pathological aspects.
Pös O, Radvanszky J, Buglyó G, Pös Z, Rusnakova D, Nagy B, Szemes T. Pös O, et al. Biomed J. 2021 Oct;44(5):548-559. doi: 10.1016/j.bj.2021.02.003. Epub 2021 Feb 13. Biomed J. 2021. PMID: 34649833 Free PMC article. Review. - Genome at juncture of early human migration: a systematic analysis of two whole genomes and thirteen exomes from Kuwaiti population subgroup of inferred Saudi Arabian tribe ancestry.
Alsmadi O, John SE, Thareja G, Hebbar P, Antony D, Behbehani K, Thanaraj TA. Alsmadi O, et al. PLoS One. 2014 Jun 4;9(6):e99069. doi: 10.1371/journal.pone.0099069. eCollection 2014. PLoS One. 2014. PMID: 24896259 Free PMC article.
References
- Feero WG, Guttmacher AE, Collins FS (2010) Genomic medicine – an updated primer. N Engl J Med 362: 2001–2011. - PubMed
- Kingsley CB (2011) Identification of causal sequence variants of disease in the next generation sequencing era. Methods Mol Biol 700: 37–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AR059781/AR/NIAMS NIH HHS/United States
- R01AR050496/AR/NIAMS NIH HHS/United States
- R01AR057049/AR/NIAMS NIH HHS/United States
- R01 AG026564/AG/NIA NIH HHS/United States
- P50AR055081/AR/NIAMS NIH HHS/United States
- R01 AR057049/AR/NIAMS NIH HHS/United States
- P50 AR055081/AR/NIAMS NIH HHS/United States
- R03 TW008221/TW/FIC NIH HHS/United States
- R01 AR050496/AR/NIAMS NIH HHS/United States
- R01AG026564/AG/NIA NIH HHS/United States
- R03TW008221/TW/FIC NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources