Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells - PubMed (original) (raw)
Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells
Ebru Okutan et al. RNA Biol. 2013 May.
Abstract
Microarray analysis of infection by a lytic Sulfolobus rudivirus, SIRV2, revealed both the temporal expression of viral genes and the differential regulation of host genes. A highly susceptible strain derived from Sulfolobus solfataricus P2 with a large genomic deletion spanning CRISPR clusters A to D was infected with SIRV2, and subjected to a microarray analysis. Transcripts from a few viral genes were detected at 15 min post-infection and all except one were expressed within 2 h. The earliest expressed genes were located mainly at the termini of the linear viral genome while later expressed genes were concentrated in the central region. Timing of the expression correlated with the known or predicted functions of the viral gene products and, thus, should facilitate functional characterization of many hypothetical viral genes. Evaluation of the microarray data with quantitative reverse-transcription PCR analyses of a few selected viral genes revealed a good correlation between the two methods. Expression of about 3,000 host genes was examined. Seventy-two were downregulated>2-fold that were mainly associated with stress response and vesicle formation, as well as chromosome structure maintenance, which appears to contribute to host chromosome degradation and cellular collapse. A further 76 host genes were upregulated>2-fold and they were dominated by genes associated with metabolism and membrane transport, including phosphate transport and DNA precursor synthesis. The altered transcriptional patterns suggest that the virus reprograms the host cellular machinery to facilitate its own DNA replication and to inhibit cellular processes required for defense against viruses.
Keywords: archaea; lytic virus; microarray; stress response; temporal regulation.
Figures
Figure 1. Susceptibility of S. solfataricus 5E6 to SIRV2 infection. (A) OD600 was measured after 1 ml fresh cells was diluted to 50 ml fresh medium with (filled squares) or without (open circles) the addition of SIRV2 (m.o.i. about 30). Cells were incubated at 78°C. (B) DNA content distributions from uninfected (left) and infected (right, m.o.i. 30) S.solfataricus 5E6 cultures were analyzed by flow cytometry time-course analysis. (C) One-step growth curve of SIRV2 infection of S. solfataricus 5E6. SIRV2 was added at an m.o.i. of about 30. The plaque forming units (PFU) are plotted against time (h p.i.)
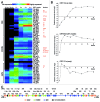
Figure 2. Temporal expression of SIRV2 genes. (A) Heat map of hierarchical clustered transcriptional changes generated from microarray data for SIRV2 genes. Genes belonging to the same group are clustered and indicated at left as early, middle and late, respectively. ORF and gp numbers for each gene are presented at the right side of the map with known or predicted functions indicated in red. hjc, Holiday junction resolvase; TMD, protein containing trans-membrane domain;sp, structure protein; pp, virus-associated pyramid protein; cp, coat protein; mt, SAM-dependent (RNA) methyltransferase; tr, putative transcription regulator. (B) qRT-PCR analysis of a selected early (ORF131a), middle (ORF56b) and late (ORF134) gene, respectively. The host RNA polymerase subunit C gene, sso0223, was equally expressed in virus infected and uninfected cells as revealed by microarray analysis and, therefore used as a reference gene. The log2 values (mean ± SD) of relative abundance of viral transcripts in relation to that of the reference gene are plotted against time (hours) p.i. (C) SIRV2 genome map showing the classification of viral genes according to results from microarray analyses (A).
Similar articles
- Protein-protein interactions leading to recruitment of the host DNA sliding clamp by the hyperthermophilic Sulfolobus islandicus rod-shaped virus 2.
Gardner AF, Bell SD, White MF, Prangishvili D, Krupovic M. Gardner AF, et al. J Virol. 2014 Jun;88(12):7105-8. doi: 10.1128/JVI.00636-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696494 Free PMC article. - Transcriptome analysis of infection of the archaeon Sulfolobus solfataricus with Sulfolobus turreted icosahedral virus.
Ortmann AC, Brumfield SK, Walther J, McInnerney K, Brouns SJ, van de Werken HJ, Bothner B, Douglas T, van de Oost J, Young MJ. Ortmann AC, et al. J Virol. 2008 May;82(10):4874-83. doi: 10.1128/JVI.02583-07. Epub 2008 Mar 12. J Virol. 2008. PMID: 18337566 Free PMC article. - Anti-CRISPR-Based and CRISPR-Based Genome Editing of Sulfolobus islandicus Rod-Shaped Virus 2.
Mayo-Muñoz D, He F, Jørgensen JB, Madsen PK, Bhoobalan-Chitty Y, Peng X. Mayo-Muñoz D, et al. Viruses. 2018 Dec 8;10(12):695. doi: 10.3390/v10120695. Viruses. 2018. PMID: 30544778 Free PMC article. - SMV1 virus-induced CRISPR spacer acquisition from the conjugative plasmid pMGB1 in Sulfolobus solfataricus P2.
Erdmann S, Shah SA, Garrett RA. Erdmann S, et al. Biochem Soc Trans. 2013 Dec;41(6):1449-58. doi: 10.1042/BST20130196. Biochem Soc Trans. 2013. PMID: 24256236 Free PMC article. Review. - Genomics and biology of Rudiviruses, a model for the study of virus-host interactions in Archaea.
Prangishvili D, Koonin EV, Krupovic M. Prangishvili D, et al. Biochem Soc Trans. 2013 Feb 1;41(1):443-50. doi: 10.1042/BST20120313. Biochem Soc Trans. 2013. PMID: 23356326 Free PMC article. Review.
Cited by
- Isolation of a virus causing a chronic infection in the archaeal model organism Haloferax volcanii reveals antiviral activities of a provirus.
Alarcón-Schumacher T, Naor A, Gophna U, Erdmann S. Alarcón-Schumacher T, et al. Proc Natl Acad Sci U S A. 2022 Aug 30;119(35):e2205037119. doi: 10.1073/pnas.2205037119. Epub 2022 Aug 22. Proc Natl Acad Sci U S A. 2022. PMID: 35994644 Free PMC article. - Anti-CRISPR proteins encoded by archaeal lytic viruses inhibit subtype I-D immunity.
He F, Bhoobalan-Chitty Y, Van LB, Kjeldsen AL, Dedola M, Makarova KS, Koonin EV, Brodersen DE, Peng X. He F, et al. Nat Microbiol. 2018 Apr;3(4):461-469. doi: 10.1038/s41564-018-0120-z. Epub 2018 Mar 5. Nat Microbiol. 2018. PMID: 29507349 Free PMC article. - Formation of a Viral Replication Focus in Sulfolobus Cells Infected by the Rudivirus Sulfolobus islandicus Rod-Shaped Virus 2.
Martínez-Alvarez L, Deng L, Peng X. Martínez-Alvarez L, et al. J Virol. 2017 Jun 9;91(13):e00486-17. doi: 10.1128/JVI.00486-17. Print 2017 Jul 1. J Virol. 2017. PMID: 28424282 Free PMC article. - Special focus CRISPR-Cas.
Marchfelder A. Marchfelder A. RNA Biol. 2013 May;10(5):655-8. doi: 10.4161/rna.24687. RNA Biol. 2013. PMID: 23872677 Free PMC article. No abstract available. - Protein-protein interactions leading to recruitment of the host DNA sliding clamp by the hyperthermophilic Sulfolobus islandicus rod-shaped virus 2.
Gardner AF, Bell SD, White MF, Prangishvili D, Krupovic M. Gardner AF, et al. J Virol. 2014 Jun;88(12):7105-8. doi: 10.1128/JVI.00636-14. Epub 2014 Apr 2. J Virol. 2014. PMID: 24696494 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources