Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein - PubMed (original) (raw)
Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein
Chris Fröhlich et al. EMBO J. 2013.
Abstract
Dynamin 1-like protein (DNM1L) mediates fission of mitochondria and peroxisomes, and dysfunction of DNM1L has been implicated in several neurological disorders. To study the molecular basis of mitochondrial remodelling, we determined the crystal structure of DNM1L that is comprised of a G domain, a bundle signalling element and a stalk. DNM1L assembled via a central stalk interface, and mutations in this interface disrupted dimerization and interfered with membrane binding and mitochondrial targeting. Two sequence stretches at the tip of the stalk were shown to be required for ordered assembly of DNM1L on membranes and its function in mitochondrial fission. In the crystals, DNM1L dimers further assembled via a second, previously undescribed, stalk interface to form a linear filament. Mutations in this interface interfered with liposome tubulation and mitochondrial remodelling. Based on these results and electron microscopy reconstructions, we propose an oligomerization mode for DNM1L which differs from that of dynamin and might be adapted to the remodelling of mitochondria.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
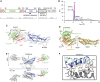
Figure 1
Structure of DNM1L. (A) Structure-based domain architecture of human DNM1L. The first and last residue of each domain is labelled as well as the modified residues for crystallization. The classical predicted domain assignment is shown below. (B) Analytical ultracentrifugation sedimentation velocity experiments for DNM1L, DNM1L ΔB-insert, DNM1L 4A and the combined mutant (4A+ΔB). The relative protein concentration c(s) as function of the normalized sedimentation coefficient _s_20,w is plotted. Dimer peaks for the DNM1L 4A and the combined mutant are indicated. Peaks in the wild-type sedimentation profile could not be assigned to oligomeric species as the protein is in a fast equilibrium between different oligomeric states. (C) Ribbon-type presentation of human DNM1L. Regions not resolved in the crystal structure, such as the B-insert, are indicated by dotted lines. Important structural elements are labelled. (D) The stalks of DNM1L molecules A and C were superimposed. A 17.5° rotation of the G domains around hinge 1 is evident. (E) Side and top view on the DNM1L dimer that assembles via the central stalk interface-2. (F) Close-up view of the dimer interface-2. Hydrogen bonds are indicated as dotted lines.
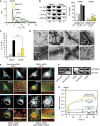
Figure 2
Dimerization via the stalk mediates oligomerization and mitochondrial remodelling. (A) Analytical ultracentrifugation sedimentation velocity experiments for DNM1L and the indicated interface-2 mutants as described in Figure 1B. Monomer and dimer peaks for E490R and K642E are indicated. As DNM1L, the E490A mutant undergoes rapid exchange reactions between different oligomeric species. (B) Left panel: Sedimentation experiments and liposome binding assays for DNM1L and DNM1L K642E. Sedimentation experiments were performed in the absence and presence of 2 mM GTP-γ-S in the absence of liposomes. Liposome co-sedimentation assays were carried out in the presence of 2 mM GDP, and in the presence or absence of PS liposomes. Lanes from SDS–PAGE are representative for three independent experiments. Boxed lanes are from the same gel. SN, supernatant. P, pellet fraction. Right panel: Quantification of sedimentation and liposome binding assays (error bars represent the s.e.m.). Bars show the percentage of protein found in the pellet with respect of total protein applied on gel. The statistical significance was calculated with respect to the corresponding DNM1L experiments. ***P<0.001; **P<0.01; *P<0.05 (also for all subsequent statistical analyses). (C) Basal and PS liposome-stimulated GTPase activities of DNM1L and the K642E mutant were determined at 37°C (_n_=2 for each experiment, error bars represent the s.e.m.). The statistical significance was calculated with respect to the corresponding DNM1L experiments. (D) Negative-stain electron microscopic analysis of DNM1L in the presence of PS liposomes and different nucleotides. The K642E mutant did not show any tubulation in the absence and presence of nucleotides. (E) Cellular localization and mitochondrial morphology studies in mito-dsRed expressing COS-7 cells. Cells depleted of DNM1L by siRNA were co-transfected with GFP, siRNA-resistant GFP-DNM1L or GFP-DNM1L K642E, respectively. Scrambled siRNA and co-transfected GFP were used as a control. Magnified boxed regions and a line scan plot with the relative fluorescence of the indicated GFP constructs and mito-dsRed are shown to the lower right of each subpanel. Scale bars: 50 μm. (F) Western blot showing efficient siRNA-mediated knock down of endogenous DNM1L. Scrambled siRNA was used as a control. Actin was stained as a loading control. Antibody efficiency was monitored using a COS cell lysate and recombinant DNM1L in a separate western blot. (G) FRAP assay for mitochondrial network connectivity. Mito-dsRed in an ROI (_d_=6 μm) containing multiple mitochondria was photobleached and its fluorescence recovery monitored for 90 s. Curves show mean values from 20 independent experiments under the indicated conditions. Prebleach intensities were normalized. For clarity, only three representative error bars are shown for each experiment. Source data for this figure is available on the online supplementary information page
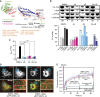
Figure 3
Functional characterization of the GPRP motif and the B-insert. (A) Ribbon-type representation of the DNM1L monomer illustrating the positions of the B-insert and the GPRP motif, the known post-translational modifications (green, black and magenta) and disease and functionally inactivating mutations (orange). The first and last visible residues of the B-insert and L2S are indicated. (B) Upper panel: Oligomerization experiments and liposome binding assays as in Figure 2B for DNM1L, DNM1L ΔB-insert, DNM1L 4A and the combined DNM1L variant (4A+ΔB). Examples are representative for three independent experiments. SN, supernatant; P, pellet fraction. Lower panel: Quantification of sedimentation and liposome binding assays. (C) Basal and PS liposome-stimulated GTPase activities of DNM1L and DNM1L mutants were determined at 37°C, as in Figure 2C. (D) Cellular localization and mitochondrial morphology studies, as in Figure 2E. COS-7 cells depleted of DNM1L by siRNA were co-transfected with mito-dsRed and siRNA-resistant DNM1L ΔB-insert and the 4A mutant. Magnified boxed regions and a line scan plot with the relative fluorescence of the indicated eGFP fusion proteins and mito-dsRed are shown at the lower right of each subpanel. Scale bars: 50 μm. (E) Mitochondrial network connectivity quantified by an FRAP assay, as in Figure 2G.
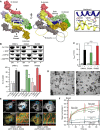
Figure 4
DNM1L oligomerizes via an alternative interface. (A) Left and middle panels: Top and side views on a surface representation of the DNM1L oligomer in the crystal. Interface-4 is indicated (white box). The direction of the oligomer is indicated by arrows. Right panel: close-up view of interface-4. Hydrogen bonds are indicated with dotted lines. (B) Oligomerization experiments and liposome binding assays for DNM1L and interface-4 mutants (E426A, R430D), as in Figure 2B. Lanes are representative of three independent experiments. SN, supernatant; P, pellet fraction. (C) Basal and PS liposome-stimulated GTPase activities of DNM1L and interface-4 mutants were determined, as in Figure 2C. (D) Representative negative-stain electron micrographs of DNM1L interface-4 mutants, as in Figure 2D. Scale bars=2 μm. (E) Cellular localization and mitochondrial morphology studies in COS-7 cells depleted of endogenous DNM1L by siRNA and co-transfected with mito-dsRed and siRNA-resistant interface-4 mutants, as in Figure 2E. Scale bars: 50 μm. (F) Mitochondrial network connectivity quantified by an FRAP assay, as in Figure 2G.
Figure 5
Structural model of the DNM1L oligomer. (A) DNM1L tetramers were manually fitted into the EM reconstruction of yeast Dnm1 (Mears et al, 2007). In this model, the stalks of DNM1L are oriented tangentially to the lipid tubule, with the B-insert pointing towards the tubule. Oligomerization of DNM1L into a filament proceeds via stalk interface-1, -2 and -3, as in dynamin and MxA. Additionally, stalk interface-4 mediates assembly of a double stalk filament. G domains could dimerize across neighbouring double filaments. (B) Schematic illustration of the DNM1L molecules in the oligomeric model. The repeating unit of the reconstruction (boxed) contains eight DNM1L monomers.
Similar articles
- A novel de novo dominant negative mutation in DNM1L impairs mitochondrial fission and presents as childhood epileptic encephalopathy.
Fahrner JA, Liu R, Perry MS, Klein J, Chan DC. Fahrner JA, et al. Am J Med Genet A. 2016 Aug;170(8):2002-11. doi: 10.1002/ajmg.a.37721. Epub 2016 May 4. Am J Med Genet A. 2016. PMID: 27145208 Free PMC article. - Functional mapping of human dynamin-1-like GTPase domain based on x-ray structure analyses.
Wenger J, Klinglmayr E, Fröhlich C, Eibl C, Gimeno A, Hessenberger M, Puehringer S, Daumke O, Goettig P. Wenger J, et al. PLoS One. 2013 Aug 19;8(8):e71835. doi: 10.1371/journal.pone.0071835. eCollection 2013. PLoS One. 2013. PMID: 23977156 Free PMC article. - DNM1L Variant Alters Baseline Mitochondrial Function and Response to Stress in a Patient with Severe Neurological Dysfunction.
Hogarth KA, Costford SR, Yoon G, Sondheimer N, Maynes JT. Hogarth KA, et al. Biochem Genet. 2018 Apr;56(1-2):56-77. doi: 10.1007/s10528-017-9829-2. Epub 2017 Nov 6. Biochem Genet. 2018. PMID: 29110115 - New insights into the function and regulation of mitochondrial fission.
Otera H, Ishihara N, Mihara K. Otera H, et al. Biochim Biophys Acta. 2013 May;1833(5):1256-68. doi: 10.1016/j.bbamcr.2013.02.002. Epub 2013 Feb 20. Biochim Biophys Acta. 2013. PMID: 23434681 Review. - Splitting up the powerhouse: structural insights into the mechanism of mitochondrial fission.
Richter V, Singh AP, Kvansakul M, Ryan MT, Osellame LD. Richter V, et al. Cell Mol Life Sci. 2015 Oct;72(19):3695-707. doi: 10.1007/s00018-015-1950-y. Epub 2015 Jun 10. Cell Mol Life Sci. 2015. PMID: 26059473 Free PMC article. Review.
Cited by
- Allosteric control of dynamin-related protein 1 through a disordered C-terminal Short Linear Motif.
Pérez-Jover I, Rochon K, Hu D, Mahajan M, Madan Mohan P, Santos-Pérez I, Ormaetxea Gisasola J, Martinez Galvez JM, Agirre J, Qi X, Mears JA, Shnyrova AV, Ramachandran R. Pérez-Jover I, et al. Nat Commun. 2024 Jan 2;15(1):52. doi: 10.1038/s41467-023-44413-6. Nat Commun. 2024. PMID: 38168038 Free PMC article. - Insights Into Mitochondrial Dynamics in Chlamydial Infection.
Yang Y, Lei W, Zhao L, Wen Y, Li Z. Yang Y, et al. Front Cell Infect Microbiol. 2022 Mar 7;12:835181. doi: 10.3389/fcimb.2022.835181. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35321312 Free PMC article. Review. - Drp1 SUMO/deSUMOylation by Senp5 isoforms influences ER tubulation and mitochondrial dynamics to regulate brain development.
Yamada S, Sato A, Ishihara N, Akiyama H, Sakakibara SI. Yamada S, et al. iScience. 2021 Dec 10;24(12):103484. doi: 10.1016/j.isci.2021.103484. eCollection 2021 Dec 17. iScience. 2021. PMID: 34988397 Free PMC article. - Role of Mitochondria in Physiology of Chondrocytes and Diseases of Osteoarthritis and Rheumatoid Arthritis.
Kan S, Duan M, Liu Y, Wang C, Xie J. Kan S, et al. Cartilage. 2021 Dec;13(2_suppl):1102S-1121S. doi: 10.1177/19476035211063858. Epub 2021 Dec 11. Cartilage. 2021. PMID: 34894777 Free PMC article. Review. - SENP3 Promotes an Mff-Primed Bcl-x L -Drp1 Interaction Involved in Cell Death Following Ischemia.
Guo C, Hildick KL, Jiang J, Zhao A, Guo W, Henley JM, Wilkinson KA. Guo C, et al. Front Cell Dev Biol. 2021 Oct 15;9:752260. doi: 10.3389/fcell.2021.752260. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34722538 Free PMC article.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 - PMC - PubMed
- Bereiter-Hahn J, Voth M, Mai S, Jendrach M (2008) Structural implications of mitochondrial dynamics. Biotechnol J 3: 765–780 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous