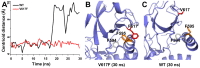Ab initio modeling and experimental assessment of Janus Kinase 2 (JAK2) kinase-pseudokinase complex structure - PubMed (original) (raw)
Ab initio modeling and experimental assessment of Janus Kinase 2 (JAK2) kinase-pseudokinase complex structure
Xiaobo Wan et al. PLoS Comput Biol. 2013 Apr.
Abstract
The Janus Kinase 2 (JAK2) plays essential roles in transmitting signals from multiple cytokine receptors, and constitutive activation of JAK2 results in hematopoietic disorders and oncogenesis. JAK2 kinase activity is negatively regulated by its pseudokinase domain (JH2), where the gain-of-function mutation V617F that causes myeloproliferative neoplasms resides. In the absence of a crystal structure of full-length JAK2, how JH2 inhibits the kinase domain (JH1), and how V617F hyperactivates JAK2 remain elusive. We modeled the JAK2 JH1-JH2 complex structure using a novel informatics-guided protein-protein docking strategy. A detailed JAK2 JH2-mediated auto-inhibition mechanism is proposed, where JH2 traps the activation loop of JH1 in an inactive conformation and blocks the movement of kinase αC helix through critical hydrophobic contacts and extensive electrostatic interactions. These stabilizing interactions are less favorable in JAK2-V617F. Notably, several predicted binding interfacial residues in JH2 were confirmed to hyperactivate JAK2 kinase activity in site-directed mutagenesis and BaF3/EpoR cell transformation studies. Although there may exist other JH2-mediated mechanisms to control JH1, our JH1-JH2 structural model represents a verifiable working hypothesis for further experimental studies to elucidate the role of JH2 in regulating JAK2 in both normal and pathological settings.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
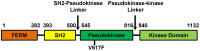
Figure 1. The domain organization of full length JAK2.
Figure 2. The schematic of hierarchical protein-protein docking procedure.
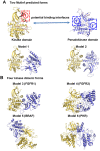
Figure 3. Starting models were generated by connecting sites identified by MutInf or by using dimer geometries from crystal structures.
(A) Two predefined packing models were constructed by manually joining the αC helix in the JAK2 kinase domain with two highly coupled sites in the JH2 pseudokinase domain identified by MutInf. (B) Four additional packing models were built by alignment to crystal structures of kinase dimeric forms (FGFR1, FGFR2, BRAF and PKB).
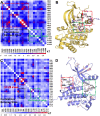
Figure 4. Correlated motions couple active to putative allosteric sites in the JAK2 JH1 and JH2 domains.
(A) The pairwise matrix of the highly coupled residues in the inactive conformation of the JAK2 JH1 domain. (B) Strong correlations between the activation loop (green box) and the αC helix (red box) are observed in the JH1 domain (yellow in cartoon). (C) The pairwise matrix of the highly coupled residues in the active conformation of the JH2 domain. (D) The strong correlations of the loop between β7–β8 sheets near the hinge region (green box) with activation loop (red box) shown in the JH2 domain (blue in cartoon).
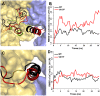
Figure 5. The V617F mutant in the JH2 domain packs closer to F595 than in the wild-type JH2 in 30 ns MD simulations.
(A) Centroid distances between F595 and V/F617 in JAK2-WT and JAK2-V617F show a closer packing in the V617F mutant than in the wild type. (B, C) Cartoon representations are shown for the last snapshot of JAK2-V617F and wild-type JH2, respectively, after 30 ns MD simulations, showing a contact in the V617F mutant that is not present after 30 ns for the wild-type pseudokinase. The F595, V/F617 and F594 are colored in orange, red and grey.
Figure 6. Motions of the activation loop and αC-helix in JAK2-WT and JAK2-V617F in 30 ns MD simulation.
(A) The JAK2-V617F mutant increases the flexibility of the activation loop in the kinase domain by restricting the movement of αC helix region in the JH2. The activation loop in the kinase domain of JAK2-WT (black) and JAK2-V617F (red) is shown alongside the crystal structure (green) in cartoon representation using PyMOL. Other residues are shown in surface representation. (B) RMSDs of Cα atoms during 30 ns of MD simulation show displacement of the kinase domain activation loop residues 994–1028 in the V617F mutant (black) but not the wildtpype (red). (C) Conformations of the αC helix in kinase domain after 30 ns MD for V617F mutant (black) and wild type (red). (D) RMSDs for the kinase αC helix (residues 586–606) in JAK2-WT and JAK2-V617F during 30 ns MD simulation show that the position of this helix is relatively stable in both cases. RMSDs are computed over Cα atoms of JAK2 with respect to the initial model after superposition of the kinase domain.
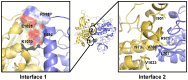
Figure 7. JAK2 JH1-JH2 complex model involves two sets of significant interfacial residues.
The JH2 (residues 523 to 816) is shown in blue cartoon while the kinase domain (residues 840 to 1132) is shown in yellow cartoon. The linker loop between two domains is colored pink. Interface 1 shows a site of electrostatic complementarity near the αC helix region of the JH2 and the αEF/αF loop region of kinase domain. The electrostatic complementarity is provided by R588, E592, E1028 and K1030. Interface 2 is composed of mainly hydrophobic residues, especially V706, L707, I901, R971, I973 and V1033.
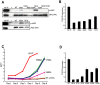
Figure 8. Mutation of interfacial residues changed JAK2 kinase activity.
(A) JAK2 mutants showed increased basal kinase activity. Activity of HA-tagged JAK2 mutants was measured by phospho-JAK2 antibodies. Total JAK2 level was measured by anti-HA antibodies. P-JAK2: phosphorylated JAK2. V: vector alone. WT: wild-type JAK2. (B) Hyperactive JAK2 mutants showed increased STAT5 activation. Activation of STAT5 was assessed using flow cytometry with Alexa647-conjugated antibodies to phospho-STAT5. Results were normalized to wild-type JAK2. (C) Hyperactive JAK2 mutants transformed BaF3/EpoR cells into factor-independent growth. Cell growth at each indicated day was measured by MTT assay. JAK2-V617F expressing cells became saturated on Day 5. WT: wild-type JAK2. (D) Mutations in JH1 reduced basal JAK2 kinase activity. Activation of STAT5 was determined as in (B). Results were normalized to wild-type JAK2.
Similar articles
- Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F.
Bandaranayake RM, Ungureanu D, Shan Y, Shaw DE, Silvennoinen O, Hubbard SR. Bandaranayake RM, et al. Nat Struct Mol Biol. 2012 Aug;19(8):754-9. doi: 10.1038/nsmb.2348. Epub 2012 Jul 22. Nat Struct Mol Biol. 2012. PMID: 22820988 Free PMC article. - Uncoupling JAK2 V617F activation from cytokine-induced signalling by modulation of JH2 αC helix.
Leroy E, Dusa A, Colau D, Motamedi A, Cahu X, Mouton C, Huang LJ, Shiau AK, Constantinescu SN. Leroy E, et al. Biochem J. 2016 Jun 1;473(11):1579-91. doi: 10.1042/BCJ20160085. Epub 2016 Mar 30. Biochem J. 2016. PMID: 27029346 Free PMC article. - New insights into the structure and function of the pseudokinase domain in JAK2.
Silvennoinen O, Ungureanu D, Niranjan Y, Hammaren H, Bandaranayake R, Hubbard SR. Silvennoinen O, et al. Biochem Soc Trans. 2013 Aug;41(4):1002-7. doi: 10.1042/BST20130005. Biochem Soc Trans. 2013. PMID: 23863170 Review. - ATP binding to the pseudokinase domain of JAK2 is critical for pathogenic activation.
Hammarén HM, Ungureanu D, Grisouard J, Skoda RC, Hubbard SR, Silvennoinen O. Hammarén HM, et al. Proc Natl Acad Sci U S A. 2015 Apr 14;112(15):4642-7. doi: 10.1073/pnas.1423201112. Epub 2015 Mar 30. Proc Natl Acad Sci U S A. 2015. PMID: 25825724 Free PMC article. - Activating Janus kinase pseudokinase domain mutations in myeloproliferative and other blood cancers.
Constantinescu SN, Leroy E, Gryshkova V, Pecquet C, Dusa A. Constantinescu SN, et al. Biochem Soc Trans. 2013 Aug;41(4):1048-54. doi: 10.1042/BST20130084. Biochem Soc Trans. 2013. PMID: 23863177 Review.
Cited by
- Molecular basis for pseudokinase-dependent autoinhibition of JAK2 tyrosine kinase.
Shan Y, Gnanasambandan K, Ungureanu D, Kim ET, Hammarén H, Yamashita K, Silvennoinen O, Shaw DE, Hubbard SR. Shan Y, et al. Nat Struct Mol Biol. 2014 Jul;21(7):579-84. doi: 10.1038/nsmb.2849. Epub 2014 Jun 11. Nat Struct Mol Biol. 2014. PMID: 24918548 Free PMC article. - JAK2 activation by growth hormone and other cytokines.
Waters MJ, Brooks AJ. Waters MJ, et al. Biochem J. 2015 Feb 15;466(1):1-11. doi: 10.1042/BJ20141293. Biochem J. 2015. PMID: 25656053 Free PMC article. Review. - Dynamic architecture of a protein kinase.
McClendon CL, Kornev AP, Gilson MK, Taylor SS. McClendon CL, et al. Proc Natl Acad Sci U S A. 2014 Oct 28;111(43):E4623-31. doi: 10.1073/pnas.1418402111. Epub 2014 Oct 15. Proc Natl Acad Sci U S A. 2014. PMID: 25319261 Free PMC article. - Role of water in the determination of protonation states of titratable residues.
Zia SR. Zia SR. J Mol Model. 2021 Jan 31;27(2):61. doi: 10.1007/s00894-021-04677-5. J Mol Model. 2021. PMID: 33517493 - Bleeding diathesis in mice lacking JAK2 in platelets.
Eaton N, Subramaniam S, Schulte ML, Drew C, Jakab D, Haberichter SL, Weiler H, Falet H. Eaton N, et al. Blood Adv. 2021 Aug 10;5(15):2969-2981. doi: 10.1182/bloodadvances.2020003032. Blood Adv. 2021. PMID: 34342643 Free PMC article.
References
- Ward AC, Touw I, Yoshimura A (2000) The Jak-Stat pathway in normal and perturbed hematopoiesis. Blood 95: 19–29. - PubMed
- Ihle JN, Gilliland DG (2007) Jak2: normal function and role in hematopoietic disorders. Curr Opin Genet Dev 17: 8–14. - PubMed
- Levine RL (2012) JAK-mutant myeloproliferative neoplasms. Curr Top Microbiol Immunol 355: 119–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous