A potential role for CHH DNA methylation in cotton fiber growth patterns - PubMed (original) (raw)
A potential role for CHH DNA methylation in cotton fiber growth patterns
Xiang Jin et al. PLoS One. 2013.
Abstract
DNA methylation controls many aspects of plant growth and development. Here, we report a novel annual growth potential change that may correlate with changes in levels of the major DNA demethylases and methyltransferases in cotton ovules harvested at different times of the year. The abundances of DNA demethylases, at both the mRNA and protein levels, increased significantly from February to August and decreased during the remainder of the 12-month period, with the opposite pattern observed for DNA methyltransferases. Over the course of one year, substantial changes in methylcytosine content was observed at certain CHH sites (H = A, C, or T) in the promoter regions of the ETHYLENE RESPONSIVE FACTOR 6 (ERF6), SUPPRESSION OF RVS 161 DELTA 4 (SUR4) and 3-KETOACYL-COA SYNTHASE 13 (KCS13), which regulate cotton fiber growth. Three independent techniques were used to confirm the annual fluctuations in DNA methylation. Furthermore, in homozygous RNAi lines specifically targeting REPRESSOR OF SILENCING 1 (ROS1, a conserved DNA demethylase domain), promotion of DNA methylation significantly reduced fiber growth during August.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
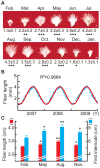
Figure 1. Annual growth potential change of cotton plants.
(A) Cotton ovule growth potential as a function of month in which ovules were harvested. Ovules were harvested 1 dpa during the month indicated, cultured for 6 d, and measured for fiber length. Numbers indicate fiber length (mean ± SE, in mm). Each ovule in this panel is a representative of thirty in the same culture. (B) Growth of cotton fibers from ovules harvested over the same monthly cycle for three consecutive years. (C) Cotton fiber length and first main stem internode length in planta in four different seasons. Error bars, SE. In (A–C), n = 6; In (A,C), *p<0.05, **p<0.01, ***p<0.001.
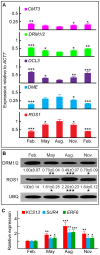
Figure 2. Annual changes in DNA methylation patterns in cotton ovules.
(A) qRT-PCR analysis of DNA methylation and demethylation genes reported in GO: 0006306. The ACT7 transcript, normalized against that of all 20 housekeeping genes reported in Table S5, was used as the internal standard. (B) Western blot analysis of DRM1/2 and ROS1. May, August, and November signal intensities were normalized to those from February (arbitrarily set to 1). Values (mean ± SE from three independent experiments) are shown beneath representative bands. Cotton UBQ was used as a loading control. (C) KCS13, SUR4 and ERF6 transcript levels changed over the course of the year, as quantified by qRT-PCR. May, August, and November values were normalized to February values (arbitrarily set to 1).
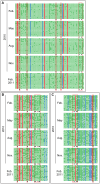
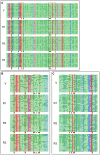
Figure 3. Bisulfite sequencing of ERF6, SUR4, and KCS13 upstream regions over one year.
The fragments to be examined were amplified by sequence-specific PCR primers after treating the template DNA with bisulfite. 17 unique “non-sister” individual clones from independent PCR reactions were selected for sequencing. Each line represents one unique “non-sister” individual bisulfite sequencing result. Only cytosines are shown using red for CG context, blue for CHG, and green for CHH. Open circles, unmethylated cytosines; closed circles, methylated cytosines; black triangles, cytosines showed annual methylation changes; red triangles, cytosine sites used for methylation-sensitive endonuclease digested PCR and Southern assay. The same designations were used for all bisulfite sequencing data reported in the current work. (A) The sequence from 272 to 662 nt in DQ464372 from ERF6 upstream region was analyzed for DNA methylation. (B) The sequence from 322 to 646 nt in JQ922563 from SUR4 upstream region was analyzed for DNA methylation. (C) The sequence from 1641 to 1939 nt in JQ922562 from KCS13 upstream region was analyzed for DNA methylation.
Figure 4. Methylation-sensitive endonuclease digested PCR and Southern analysis of ERF6, SUR4, and KCS13 upstream regions over one year.
(A) Methylation-sensitive endonuclease digested PCR amplification of ERF6 upstream region. Top: schematic diagram of the identification of a methylation-sensitive _BstX_I digenstion site (CCANNNNNNTGG) at −275 bp of the ERF6 promoter. The bold C indicates a CHH site with annual methylation pattern change, corresponding to the cytosine labelled with red triangles in Figure 3A. Bottom: PCR amplification using genomic DNA with (+) or without (−) _BstX_I digestion. (B) Southern blot of genomic DNA harvested at different times of the year, first digested by a methylation non-sensitive endonuclease _Mbo_II (TCTTC) to obtain a full length fragment of 605 bp from −621 to −15 of ERF6 upstream regions, then digested thoroughly with _BstX_I, and probed with the fragment from −263 to −21 nt. The signal intensities of the band of _BstX_I-cleaved 244 bp changed at different time-of-year (see Table S8), indicating the methlytion levels of this CHH site were different, consistent with the bisulfite sequencing data in Figure 3A and methylation-sensitive endonuclease digested PCR results in Figure 4A. The same methylation-sensitive endonuclease digested PCR experiments were performed for the upstream regions of SUR4 (C) and KCS13 (E), except the methylation-sensitive endonucleases used were _HinF_I and _Bsl_I, respectively. Further, the same methylation-sensitive endonuclease digested Southern experiments were performed for the upstream regions of SUR4 (D) and KCS13 (F), except the genomic DNA were first digested by _Bcl_I (TGATCA) and _NSi_I (ATGCAT), then digested by methylation-sensitive endonucleases _HinF_I (GANTC) and _Bsl_I (CCNNNNNNNGG), respectively. The signal intensities of _HinF_I- and _Bsl_I-cleaved 330 bp and 837 bp changed similarly (see Table S8).
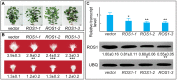
Figure 5. Phenotype and genetic identification of ROS1RNAi lines.
(A) Homozygous ROS1RNAi cotton lines at flowering. Vector plants carry the empty vector and showed identical properties with the parent. (B) Cotton ovules from RNAi lines that flowered during August (upper panel) or February (lower panel) were cultured for 6 d before being photographed for fiber measurement. (C) Analysis of ROS1 transcripts in ovules from various ROS1RNAi lines by qRT-PCR (upper panel) and western blotting (lower panel).
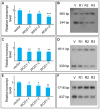
Figure 6. Bisulfite sequencing of ERF6, SUR4, and KCS13 upstream regions in ROS1RNAi lines.
The same primers were used for bisulfite treated PCR and sequencing as in Figure 3. (A), promoter region of ERF6; (B), promoter region of SUR4; (C), promoter region of KCS13; V, RNAi line with empty vector; R1–R3, RNAi line ROS1-1 to ROS1-3. All symbols are same to Figure 3.
Figure 7. Methylation-sensitive endonuclease digested PCR and Southern analysis of ERF6, SUR4, and KCS13 upstream regions in ROS1RNAi lines.
(A) Analysis of relative ERF6 transcription in ovules from ROS1RNAi lines by qRT-PCR. The level of ERF6 transcripts in ovules from the empty vector line (V) was arbitrarily defined as 1. (B) Southern blot analysis of genomic DNA prepared from ROS1RNAi lines digested thoroughly with _BstX_I. See detailed information in Figure 4 legend. Similar qRT-PCR experiments were performed for SUR4 (C) and KCS13 (E) transcriptions, as well as similar Southern experiments for SUR4 (D) and KCS13 (F), respectively. Note the reduced intensities of the _BstX_I-, _HinF_I- and _Bsl_I-cleaved bands in all three RNAi lines compared to the vector line.
Similar articles
- Dynamic Roles for Small RNAs and DNA Methylation during Ovule and Fiber Development in Allotetraploid Cotton.
Song Q, Guan X, Chen ZJ. Song Q, et al. PLoS Genet. 2015 Dec 28;11(12):e1005724. doi: 10.1371/journal.pgen.1005724. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26710171 Free PMC article. - Phytohormonal networks promote differentiation of fiber initials on pre-anthesis cotton ovules grown in vitro and in planta.
Kim HJ, Hinchliffe DJ, Triplett BA, Chen ZJ, Stelly DM, Yeater KM, Moon HS, Gilbert MK, Thyssen GN, Turley RB, Fang DD. Kim HJ, et al. PLoS One. 2015 Apr 30;10(4):e0125046. doi: 10.1371/journal.pone.0125046. eCollection 2015. PLoS One. 2015. PMID: 25927364 Free PMC article. - A comparative miRNAome analysis reveals seven fiber initiation-related and 36 novel miRNAs in developing cotton ovules.
Wang ZM, Xue W, Dong CJ, Jin LG, Bian SM, Wang C, Wu XY, Liu JY. Wang ZM, et al. Mol Plant. 2012 Jul;5(4):889-900. doi: 10.1093/mp/ssr094. Epub 2011 Dec 1. Mol Plant. 2012. PMID: 22138860 - Polyploidy and small RNA regulation of cotton fiber development.
Guan X, Song Q, Chen ZJ. Guan X, et al. Trends Plant Sci. 2014 Aug;19(8):516-28. doi: 10.1016/j.tplants.2014.04.007. Epub 2014 May 25. Trends Plant Sci. 2014. PMID: 24866591 Review. - Gene expression changes and early events in cotton fibre development.
Lee JJ, Woodward AW, Chen ZJ. Lee JJ, et al. Ann Bot. 2007 Dec;100(7):1391-401. doi: 10.1093/aob/mcm232. Epub 2007 Sep 27. Ann Bot. 2007. PMID: 17905721 Free PMC article. Review.
Cited by
- Interactive roles of chromatin regulation and circadian clock function in plants.
Chen ZJ, Mas P. Chen ZJ, et al. Genome Biol. 2019 Mar 22;20(1):62. doi: 10.1186/s13059-019-1672-9. Genome Biol. 2019. PMID: 30902105 Free PMC article. Review. - Global analysis of DNA methylation in young (J1) and senescent (J2) Gossypium hirsutum L. cotyledons by MeDIP-Seq.
Dou L, Jia X, Wei H, Fan S, Wang H, Guo Y, Duan S, Pang C, Yu S. Dou L, et al. PLoS One. 2017 Jul 17;12(7):e0179141. doi: 10.1371/journal.pone.0179141. eCollection 2017. PLoS One. 2017. PMID: 28715427 Free PMC article. - Analyses of Methylomes Derived from Meso-American Common Bean (Phaseolus vulgaris L.) Using MeDIP-Seq and Whole Genome Sodium Bisulfite-Sequencing.
Crampton M, Sripathi VR, Hossain K, Kalavacharla V. Crampton M, et al. Front Plant Sci. 2016 Apr 26;7:447. doi: 10.3389/fpls.2016.00447. eCollection 2016. Front Plant Sci. 2016. PMID: 27199997 Free PMC article. - Recent insights into cotton functional genomics: progress and future perspectives.
Ashraf J, Zuo D, Wang Q, Malik W, Zhang Y, Abid MA, Cheng H, Yang Q, Song G. Ashraf J, et al. Plant Biotechnol J. 2018 Mar;16(3):699-713. doi: 10.1111/pbi.12856. Epub 2018 Jan 15. Plant Biotechnol J. 2018. PMID: 29087016 Free PMC article. Review. - Multi-omics maps of cotton fibre reveal epigenetic basis for staged single-cell differentiation.
Wang M, Wang P, Tu L, Zhu S, Zhang L, Li Z, Zhang Q, Yuan D, Zhang X. Wang M, et al. Nucleic Acids Res. 2016 May 19;44(9):4067-79. doi: 10.1093/nar/gkw238. Epub 2016 Apr 11. Nucleic Acids Res. 2016. PMID: 27067544 Free PMC article.
References
- Harmer SL, Panda S, Kay SA (2001) Molecular bases of circadian rhythms. Annu Rev Cell Biol 17: 215–253. - PubMed
- Doi M, Hirayama J, Sassone-Corsi P (2006) Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508. - PubMed
Publication types
MeSH terms
Grants and funding
This work was supported by grants from the China National Basic Research Program (Grant number 2010CB126002), the National Natural Science Foundation of China (Grant number 90717009), and the 111 Project funded by the Chinese Ministry of Education. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials