Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation - PubMed (original) (raw)
Comparative Study
. 2013 Apr 11;8(4):e60549.
doi: 10.1371/journal.pone.0060549. Print 2013.
Zhiping Yao, Rishi Arora, Sachin Thigale, Ieuan Clay, Bruno Inverardi, Joy Fletcher, Paul Taslimi, Michael G Acker, Bertran Gerrits, Johannes Voshol, Andreas Bauer, Dirk Schübeler, Tewis Bouwmeester, Heinz Ruffner
Affiliations
- PMID: 23593242
- PMCID: PMC3623819
- DOI: 10.1371/journal.pone.0060549
Comparative Study
Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation
Michael Brauchle et al. PLoS One. 2013.
Abstract
Histone modifications play an important role in chromatin organization and gene regulation, and their interpretation is referred to as epigenetic control. The methylation levels of several lysine residues in histone tails are tightly controlled, and JmjC domain-containing proteins are one class of broadly expressed enzymes catalyzing methyl group removal. However, several JmjC proteins remain uncharacterized, gaps persist in understanding substrate recognition, and the integration of JmjC proteins into signaling pathways is just emerging. The KDM3 subfamily is an evolutionarily conserved group of histone demethylase proteins, thought to share lysine substrate specificity. Here we use a systematic approach to compare KDM3 subfamily members. We show that full-length KDM3A and KDM3B are H3K9me1/2 histone demethylases whereas we fail to observe histone demethylase activity for JMJD1C using immunocytochemical and biochemical approaches. Structure-function analyses revealed the importance of a single amino acid in KDM3A implicated in the catalytic activity towards H3K9me1/2 that is not conserved in JMJD1C. Moreover, we use quantitative proteomic analyses to identify subsets of the interactomes of the 3 proteins. Specific interactor candidates were identified for each of the three KDM3 subfamily members. Importantly, we find that SCAI, a known transcriptional repressor, interacts specifically with KDM3B. Taken together, we identify substantial differences in the biology of KDM3 histone demethylases, namely enzymatic activity and protein-protein interactions. Such comparative approaches pave the way to a better understanding of histone demethylase specificity and protein function at a systems level and are instrumental in identifying the more subtle differences between closely related proteins.
Conflict of interest statement
Competing Interests: Michael Brauchle, Zhiping Yao, Rishi Arora, Sachin Thigale, Ieuan Clay, Bruno Inverardi, Joy Fletcher, Paul Taslimi, Michael G. Acker, Bertran Gerrits, Johannes Voshol, Andreas Bauer, Tewis Bouwmeester and Heinz Ruffner are all employees of Novartis AG. Dirk Schübeler is an employee of FMI, largely funded by Novartis AG. This does not alter the authors' adherence to all the PLOS ONE policies on sharing data and materials.
Figures
Figure 1. Enzymatic activity of KDM3 subfamily members towards H3K9 methylation.
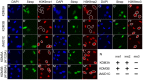
Individual KDM3 subfamily members were transiently overexpressed in U-2 OS cells. (A-M) DAPI staining indicating cell nuclei. (A'-M') Cellular expression of Avi-tagged KDM3 subfamily members, as detected by streptavidin-AlexaFluor-488 recognizing the biotinylated Avi-tag. (A’’-M’’) H3K9me1, -me2 or -me3 groups, respectively, as detected by antibody staining. White circles outline the transfected cells in the last panel of each series. Note that cells transfected with KDM3A and KDM3B (A, D, G and B, E, H) abolish H3K9me1 (A’’ and B’’) and -me2 (D’’ and E’’) but not -me3 (G’’ and H’’) staining. On the other hand, JMJD1C transfection (C, F, I) does not decrease H3K9me1 (C’’), -me2 (F’’) or -me3 (I’’) levels. The catalytic mutant versions of KDM3A(H1120A) (J, L) and KDM3B(H1560A) (K, M) neither reduce H3Kme1 (J’’, K’’) nor H3K9me2 (L’’, M’’) levels. N shows the summary of the enzymatic activity described above.
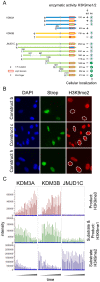
Figure 2. Domain mapping of KDM3 subfamily members identifies regions important for demethylase activity towards methylated H3K9.
(A) Overview of constructs used in this study (left) and summary of results obtained for each construct with regard to demethylase activity towards H3K9 and subcellular localization (right). Full-length and truncated KDM3A (a and d, respectively) and full-length KDM3B (e) show activity towards H3K9me1 and -me2. Full-length and truncated versions of JMJD1C (g and i-o, respectively) do not show any enzymatic activity against either H3K9me1 or -me2. Construct i corresponds to the alternative splice isoform 2 of JMJD1C. Note that constructs d and m as well as e and j are similar in size, respectively. The star denotes the Y to F mutation in KDM3B (f), the red box denotes the JmjC domain in each construct, the grey box denotes the putative Zinc finger. (B) Hybrid constructs in which the JmjC domain in KDM3A was exchanged with the one of KDM3B (Construct b) or JMJD1C (Construct c) were assayed for their ability to demethylate H3K9me2 and –me1. Whereas construct b was active against both –me2 and –me1, construct c was inactive against both methyl groups. The hybrid construct in which the JmjC domain in JMJD1C was exchanged with the one of KDM3A (Construct h) can neither remove methyl group H3K9me2 nor –me1; only the data for –me2 are shown for either construct. (C) MS-based assessment of KMD3A, KDM3B and JMJD1C catalytic activity towards H3K9me2 and –me1. H3K9me2 peptides were incubated for 2 hours with the required co-factors and either recombinant KDM3A (aa511-1321), KDM3B(aa879-1761) or JMJD1C (aa1696–2540). Along H3K9me2 substrate, H3K9me1 and H3K9me0 reaction products were quantified using MS. Reactions were performed in triplicates, and H3K9me0, –me1 and –me2 levels were measured at 7 time intervals during the 2 hour incubation period, hence the 21 peaks shown per sample. Note that in the case of KDM3A and KDM3B, H3K9me2 levels strongly and H3K9me1 levels weakly drop during the incubation period, while H3K9me0 levels steadily increase over the course of the experiment. Using JMJD1C, neither H3K9me0 nor –me1 were produced over time up to the end of the 2 hour incubation period, indicating that JMJD1C cannot demethylate H3K9me1 or –me2.
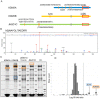
Figure 3. MS analysis of KDM3 subfamily members.
(A) Phosphorylated peptides and residues identified in overexpressed KDM3A, KDM3B and JMJD1C using MS. Amino acid positions of the phosphorylated sites are indicated in the respective protein. Underlined phosphorylated sites are conserved. Potential phospho-sites within identified phospho-peptides are indicated in italics and brackets. (B) MS/MS spectra of the KDM3B peptide containing phosphorylated Y1541 (underlined). (C) Coomassie-stained gels showing affinity purifications of Avi-tagged, overexpressed KDM3 subfamily members from lysates of transfected HEK293T cells. The different lanes show individual purifications of KDM3A, KDM3B, JMJD1C C-term and JMJD1C as well as a control purification from mock-transfected HEK293T cells. The positions of the individually overexpressed proteins are indicated by orange squares, the position of the KDM3B interactor SCAI is indicated by a blue square. These samples were subjected to quantitative MS analysis. (D) Relative enrichment of KDM3B interactor candidates in relation to the mock control. The 406 proteins identified with at least 4 peptides were binned into 45 columns; stippled lines indicate 2 standard deviations from the mean. Proteins that centered around 0 were not enriched, whereas proteins retrieved on KDM3B that were enriched with ≥2 standard deviations (right stippled line) were considered KDM3B candidate interactors. KDM3B and its interactor candidate SCAI are indicated by arrows and boxed in the same color as in C.
Figure 4. The Zinc finger mutant KDM3A T667A loses its ability to efficiently demethylate H3K9me1.
(A) Sequence alignment of the three zinc finger domains of the KDM3 subfamily members. Amino acids marked in red are fully conserved between all three proteins, amino acids marked in orange are conserved in KDM3B and JMJD1C but not in KDM3A, and amino acids marked in white are conserved between KDM3A and KMD3B but not JMJD1C. The latter served as template to convert each amino acid in KDM3A to the corresponding amino acid present in the JMJD1C zinc finger domain, as indicated in green. (B) The four zinc finger mutants generated in KMD3A were analyzed for their ability to demethylate H3K9me1 and –me2 using a biochemical approach combined with a MS-based readout, similarly as described in Fig. 2C. KDM3A T667A revealed reduced activity towards H3K9me2 and strongly reduced activity towards H3K9me1 under the conditions tested. The other three zinc finger mutants behaved like wild-type KDM3A. (C) The same four zinc finger mutants were analyzed upon transient overexpression as GFP-NLS-fusion proteins in HEK293T cells for their ability to demethylate H3K9me1 and –me2. The following constructs were employed: a-c, n-p: GFP-NLS-KDM3A-V664A; d-f, q-s: GFP-NLS-KDM3A-T667A; g-i, t-v: GFP-NLS-KDM3A-P677Q; k-m, w-y: GFP-NLS-KDM3A-G682V. Lanes a,d,g,k and n,q,t,w: DAPI; lanes b,e,h,l and o,r,u,x: GFP to monitor transfected cells; lanes c,f,i,m and p,s,v,y: methylation state of H3K9me1 and -me2, respectively. GFP-NLS-KDM3A-T667A lacks activity against H3K9me1 but retains activity against H3K9me2 (f and s), while the other three mutants are active against both H3K9me1 and –me2 (c,i,m and p,v,y).
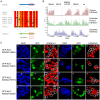
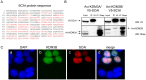
Figure 5. SCAI is a specific interactor candidate of KDM3B.
(A) SCAI protein sequence with the peptides identified by MS highlighted in red. The amino acids marked in green indicate trypsin cleavage sites. SCAI sequence coverage by MS was 51%. (B) Reciprocal co-immunoprecipitation of SCAI and KDM3B. V5-SCAI was either co-expressed with Avi-KDM3A or Avi-KDM3B. Reciprocal co-immunoprecipitations using V5- antibodies or streptavidin-coated beads were performed and the immunoprecipitated proteins from each immunoprecipitation were separated on SDS gels. A V5-antibody and streptavidin-HRP were used to detect SCAI and KDM3A or KDM3B, respectively. Only KDM3B but not KDM3A co-precipitated with and was able to precipitate V5-SCAI, respectively. (C) Sub-cellular co-localization of KDM3B and SCAI in HEK293T cells. Avi-KDM3B and V5-SCAI were co-expressed in HEK293T cells and detected by immunoreagents against their respective tags (b and c). The two proteins were found to co-localize in the nucleus (d).
Similar articles
- Histone Demethylase KDM3 (JMJD1) in Transcriptional Regulation and Cancer Progression.
Fan L, Sudeep K, Qi J. Fan L, et al. Adv Exp Med Biol. 2023;1433:69-86. doi: 10.1007/978-3-031-38176-8_4. Adv Exp Med Biol. 2023. PMID: 37751136 Free PMC article. - Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen regulates the KSHV epigenome by association with the histone demethylase KDM3A.
Kim KY, Huerta SB, Izumiya C, Wang DH, Martinez A, Shevchenko B, Kung HJ, Campbell M, Izumiya Y. Kim KY, et al. J Virol. 2013 Jun;87(12):6782-93. doi: 10.1128/JVI.00011-13. Epub 2013 Apr 10. J Virol. 2013. PMID: 23576503 Free PMC article. - The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36.
Klose RJ, Yamane K, Bae Y, Zhang D, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. Klose RJ, et al. Nature. 2006 Jul 20;442(7100):312-6. doi: 10.1038/nature04853. Epub 2006 May 28. Nature. 2006. PMID: 16732292 - Advances in Histone Demethylase KDM3A as a Cancer Therapeutic Target.
Yoo J, Jeon YH, Cho HY, Lee SW, Kim GW, Lee DH, Kwon SH. Yoo J, et al. Cancers (Basel). 2020 Apr 28;12(5):1098. doi: 10.3390/cancers12051098. Cancers (Basel). 2020. PMID: 32354028 Free PMC article. Review. - Emerging Roles of JmjC Domain-Containing Proteins.
Accari SL, Fisher PR. Accari SL, et al. Int Rev Cell Mol Biol. 2015;319:165-220. doi: 10.1016/bs.ircmb.2015.07.003. Epub 2015 Aug 19. Int Rev Cell Mol Biol. 2015. PMID: 26404469 Review.
Cited by
- KDM3A coordinates actin dynamics with intraflagellar transport to regulate cilia stability.
Yeyati PL, Schiller R, Mali G, Kasioulis I, Kawamura A, Adams IR, Playfoot C, Gilbert N, van Heyningen V, Wills J, von Kriegsheim A, Finch A, Sakai J, Schofield CJ, Jackson IJ, Mill P. Yeyati PL, et al. J Cell Biol. 2017 Apr 3;216(4):999-1013. doi: 10.1083/jcb.201607032. Epub 2017 Feb 28. J Cell Biol. 2017. PMID: 28246120 Free PMC article. - Epigenetic roles of KDM3B and KDM3C in tumorigenesis and their therapeutic implications.
Yoo J, Kim GW, Jeon YH, Lee SW, Kwon SH. Yoo J, et al. Cell Death Dis. 2024 Jun 26;15(6):451. doi: 10.1038/s41419-024-06850-z. Cell Death Dis. 2024. PMID: 38926399 Free PMC article. Review. - The Arabidopsis JMJ29 Protein Controls Circadian Oscillation through Diurnal Histone Demethylation at the CCA1 and PRR9 Loci.
Lee HG, Seo PJ. Lee HG, et al. Genes (Basel). 2021 Apr 5;12(4):529. doi: 10.3390/genes12040529. Genes (Basel). 2021. PMID: 33916408 Free PMC article. - KDM3A-mediated SP1 activates PFKFB4 transcription to promote aerobic glycolysis in osteosarcoma and augment tumor development.
Wang W, Wang B. Wang W, et al. BMC Cancer. 2022 May 19;22(1):562. doi: 10.1186/s12885-022-09636-8. BMC Cancer. 2022. PMID: 35590288 Free PMC article. - KDM3B suppresses APL progression by restricting chromatin accessibility and facilitating the ATRA-mediated degradation of PML/RARα.
Wang X, Fan H, Xu C, Jiang G, Wang H, Zhang J. Wang X, et al. Cancer Cell Int. 2019 Oct 4;19:256. doi: 10.1186/s12935-019-0979-7. eCollection 2019. Cancer Cell Int. 2019. PMID: 31592194 Free PMC article.
References
- Campos EI, Reinberg D (2009) Histones: Annotating Chromatin. Annu Rev Genet 43: 559–599. - PubMed
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705. - PubMed
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
The authors have no support or funding to report.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases