Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer - PubMed (original) (raw)
. 2013 Apr 17;5(181):181re1.
doi: 10.1126/scitranslmed.3006112.
Erhan Bilal, Erich Huang, Thea C Norman, Lars Ottestad, Brigham H Mecham, Ben Sauerwine, Michael R Kellen, Lara M Mangravite, Matthew D Furia, Hans Kristian Moen Vollan, Oscar M Rueda, Justin Guinney, Nicole A Deflaux, Bruce Hoff, Xavier Schildwachter, Hege G Russnes, Daehoon Park, Veronica O Vang, Tyler Pirtle, Lamia Youseff, Craig Citro, Christina Curtis, Vessela N Kristensen, Joseph Hellerstein, Stephen H Friend, Gustavo Stolovitzky, Samuel Aparicio, Carlos Caldas, Anne-Lise Børresen-Dale
Affiliations
- PMID: 23596205
- PMCID: PMC3897241
- DOI: 10.1126/scitranslmed.3006112
Systematic analysis of challenge-driven improvements in molecular prognostic models for breast cancer
Adam A Margolin et al. Sci Transl Med. 2013.
Abstract
Although molecular prognostics in breast cancer are among the most successful examples of translating genomic analysis to clinical applications, optimal approaches to breast cancer clinical risk prediction remain controversial. The Sage Bionetworks-DREAM Breast Cancer Prognosis Challenge (BCC) is a crowdsourced research study for breast cancer prognostic modeling using genome-scale data. The BCC provided a community of data analysts with a common platform for data access and blinded evaluation of model accuracy in predicting breast cancer survival on the basis of gene expression data, copy number data, and clinical covariates. This approach offered the opportunity to assess whether a crowdsourced community Challenge would generate models of breast cancer prognosis commensurate with or exceeding current best-in-class approaches. The BCC comprised multiple rounds of blinded evaluations on held-out portions of data on 1981 patients, resulting in more than 1400 models submitted as open source code. Participants then retrained their models on the full data set of 1981 samples and submitted up to five models for validation in a newly generated data set of 184 breast cancer patients. Analysis of the BCC results suggests that the best-performing modeling strategy outperformed previously reported methods in blinded evaluations; model performance was consistent across several independent evaluations; and aggregating community-developed models achieved performance on par with the best-performing individual models.
Figures
Fig. 1. Timeline and phases of BCC
At initiation (phase 1), a subset of the METABRIC data set was provided along with orientation on how to use the Synapse platform for accessing data and submitting models and source code. Phase 2 provided a new randomization of samples, to eliminate biases in the distribution of clinical variables across training and test data, and renormalization of METABRIC mRNA expression and DNA copy number data, to reduce batch effects and harmonize data with the OsloVal data used in phase 3. During phase 2, there was a live “pre–15 October 2012” leaderboard that provided real-time scores for each submission against the held-out test set of 500 samples. At the conclusion of phase 2 on 15 October 2012, all models in the leaderboard were tested against the remaining held-out 481 samples. In the final validation round (phase 3), participants were invited to retrain up to five models on the entire METABRIC data set. Each model was then assigned a final CI score and consequently a rank based on the model’s performance against the independent OsloVal test set.
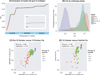
Fig. 2. BCC through time
(A) During phase 2, the highest-scoring model scores were recorded for each date until the leaderboard was closed. Each colored segment represents a top-scoring team at any given point for the period extending from late September 2012 until the final deadline of phase 2 (15 October 2012). The plot records only the times when there was an increase in the best score, whereas the teams that achieved this score are labeled with different colors. The sequence of colors highlights an important aspect of the real-time feedback, where teams were encouraged to improve their models after being bested on the leaderboard by another team. Inset, the same plot with a y-axis scale ranging from of 0.5 to 1.0 maximum CI. (B) Probability density function plots of model scores posted on the live pre–15 October leaderboard evaluated against (i) the first test set of 500 samples (blue), (ii) the second test set of 481 samples (yellow), and (iii) the OsloVal data set (red). The null hypothesis probability density, which corresponds to random predictions evaluated against the OsloVal data set, is shown in purple. (C) Scatter plot of pre–15 October 2012 model performance versus 15 October 2012 performance. Colors represent quantiles, meaning that the ordered data are divided into four equal groups numbered consecutively from the bottom-scoring models (1) to the top-scoring models (4) for pre–15 October model performance. (D) Scatter plot of pre–15 October 2012 model performance versus final OsloVal performance. Colors represent quantiles of pre–15 October model performance. Asterisk represents the highest-scoring submitted model.
Fig. 3. Rank stability of final models
The OsloVal test data were randomly subsampled 100 times using 80% of the samples. Model rank was recalculated at each iteration. Models are ordered by their final posted leaderboard score (P values for the top three models, which were submitted by the same team, versus the fourth place model were as follows = 5.1 × 10−28, 1.8 × 10−22, and 1.7 × 10−20 by Wilcoxon rank-sum tests). With these box plots, the middle horizontal line represents the median, the upper whisker extends to the highest value within a 1.5× distance between the first and third quantiles, and the lower whisker extends to the lowest value within a 1.5× distance. Data beyond the ends of the whiskers are outliers plotted as points.
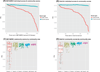
Fig. 4. Individual and community scores for METABRIC and OsloVal
(A) Individual model scores are ordered by their rank on the pre–15 October 2012 METABRIC leaderboard (red line). For each model rank (displayed on the x axis), the blue line plots the aggregate model score based on combining all models less than or equal to the given rank. (B) Individual and aggregate model scores based on evaluation in the OsloVal data set. (C) Individual model scores (that is, community = 1) from the pre–15 October 2012 METABRIC leaderboard (
http://leaderboards.bcc.sagebase.org/pre\_oct15/index.html
) are plotted alongside the community aggregate scores obtained when 5, 10, 20, and 50 randomly chosen models were considered. (D) Individual model scores (that is, community = 1) from the final OsloVal leaderboard (
http://leaderboards.bcc.sagebase.org/final/index.html
) are plotted alongside the community aggregate scores obtained when 5, 10, 20, and 50 randomly chosen predictions were considered. The colors correspond to community size: red = 1, yellow = 5, green = 10, blue = 20, purple = 50.
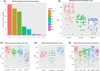
Fig. 5. Model performance and clinical characteristics
(A) Percentage of CI variance explained by each clinical variable. (B) CIs were calculated for OsloVal models according to subsets of patients by histological grade. (C) CIs were calculated for OsloVal models according to subsets of patients by LN status. (D) CIs were calculated for OsloVal models according to subsets of patients by follow-up time (OS). (E) CIs were calculated for OsloVal models according to subsets of patients by age, ER status, and HER2 status. Patients were divided into subsets according to each of the above clinical characteristics. Individual model predictions were generated for patients belonging to each subset, and the CI was calculated by comparison with the actual survival for each patient.
Comment in
- Prognostic models: rising to the challenge.
McCarthy N. McCarthy N. Nat Rev Cancer. 2013 Jun;13(6):378. doi: 10.1038/nrc3530. Epub 2013 May 3. Nat Rev Cancer. 2013. PMID: 23640208 No abstract available.
Similar articles
- Development of a prognostic model for breast cancer survival in an open challenge environment.
Cheng WY, Ou Yang TH, Anastassiou D. Cheng WY, et al. Sci Transl Med. 2013 Apr 17;5(181):181ra50. doi: 10.1126/scitranslmed.3005974. Sci Transl Med. 2013. PMID: 23596202 - Mixture classification model based on clinical markers for breast cancer prognosis.
Zeng T, Liu J. Zeng T, et al. Artif Intell Med. 2010 Feb-Mar;48(2-3):129-37. doi: 10.1016/j.artmed.2009.07.008. Epub 2009 Dec 14. Artif Intell Med. 2010. PMID: 20005686 - Identification of a 6-gene signature for the survival prediction of breast cancer patients based on integrated multi-omics data analysis.
Mo W, Ding Y, Zhao S, Zou D, Ding X. Mo W, et al. PLoS One. 2020 Nov 10;15(11):e0241924. doi: 10.1371/journal.pone.0241924. eCollection 2020. PLoS One. 2020. PMID: 33170908 Free PMC article. - Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.
Williams C, Brunskill S, Altman D, Briggs A, Campbell H, Clarke M, Glanville J, Gray A, Harris A, Johnston K, Lodge M. Williams C, et al. Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170 Review. - The clinical effectiveness and cost-effectiveness of different surveillance mammography regimens after the treatment for primary breast cancer: systematic reviews registry database analyses and economic evaluation.
Robertson C, Arcot Ragupathy SK, Boachie C, Dixon JM, Fraser C, Hernández R, Heys S, Jack W, Kerr GR, Lawrence G, MacLennan G, Maxwell A, McGregor J, Mowatt G, Pinder S, Ternent L, Thomas RE, Vale L, Wilson R, Zhu S, Gilbert FJ. Robertson C, et al. Health Technol Assess. 2011 Sep;15(34):v-vi, 1-322. doi: 10.3310/hta15340. Health Technol Assess. 2011. PMID: 21951942 Free PMC article. Review.
Cited by
- Prediction of response to anti-cancer drugs becomes robust via network integration of molecular data.
Franco M, Jeggari A, Peuget S, Böttger F, Selivanova G, Alexeyenko A. Franco M, et al. Sci Rep. 2019 Feb 20;9(1):2379. doi: 10.1038/s41598-019-39019-2. Sci Rep. 2019. PMID: 30787419 Free PMC article. - A crowd-sourcing approach for the construction of species-specific cell signaling networks.
Bilal E, Sakellaropoulos T, Melas IN, Messinis DE, Belcastro V, Rhrissorrakrai K, Meyer P, Norel R, Iskandar A, Blaese E, Rice JJ, Peitsch MC, Hoeng J, Stolovitzky G, Alexopoulos LG, Poussin C; Challenge Participants. Bilal E, et al. Bioinformatics. 2015 Feb 15;31(4):484-91. doi: 10.1093/bioinformatics/btu659. Epub 2014 Oct 7. Bioinformatics. 2015. PMID: 25294919 Free PMC article. - Immunohistochemical Evaluation of FGD3 Expression: A New Strong Prognostic Factor in Invasive Breast Cancer.
Susini T, Saccardin G, Renda I, Giani M, Tartarotti E, Nori J, Vanzi E, Pasqualini E, Bianchi S. Susini T, et al. Cancers (Basel). 2021 Jul 29;13(15):3824. doi: 10.3390/cancers13153824. Cancers (Basel). 2021. PMID: 34359725 Free PMC article. - Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data.
Jang IS, Neto EC, Guinney J, Friend SH, Margolin AA. Jang IS, et al. Pac Symp Biocomput. 2014:63-74. Pac Symp Biocomput. 2014. PMID: 24297534 Free PMC article. - Innovation and cumulative culture through tweaks and leaps in online programming contests.
Miu E, Gulley N, Laland KN, Rendell L. Miu E, et al. Nat Commun. 2018 Jun 13;9(1):2321. doi: 10.1038/s41467-018-04494-0. Nat Commun. 2018. PMID: 29899424 Free PMC article.
References
- Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer. 2013;132:1133–1145. - PubMed
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, Pergamenschikov A, Williams C, Zhu SX, Lønning PE, Børresen-Dale AL, Brown PO, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. - PubMed
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, Rijn Mvan de S.S. Jeffrey, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10869–10874. - PMC - PubMed
- Vijver van de MJ, He YD, van’t Veer LJ, Dai H, Hart AAM, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. - PubMed
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004;351:2817–2826. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical