Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide - PubMed (original) (raw)
. 2013 May 2;497(7447):137-41.
doi: 10.1038/nature12120. Epub 2013 Apr 21.
Aashish Manglik, Andrew C Kruse, Kunhong Xiao, Rosana I Reis, Wei-Chou Tseng, Dean P Staus, Daniel Hilger, Serdar Uysal, Li-Yin Huang, Marcin Paduch, Prachi Tripathi-Shukla, Akiko Koide, Shohei Koide, William I Weis, Anthony A Kossiakoff, Brian K Kobilka, Robert J Lefkowitz
Affiliations
- PMID: 23604254
- PMCID: PMC3654799
- DOI: 10.1038/nature12120
Structure of active β-arrestin-1 bound to a G-protein-coupled receptor phosphopeptide
Arun K Shukla et al. Nature. 2013.
Abstract
The functions of G-protein-coupled receptors (GPCRs) are primarily mediated and modulated by three families of proteins: the heterotrimeric G proteins, the G-protein-coupled receptor kinases (GRKs) and the arrestins. G proteins mediate activation of second-messenger-generating enzymes and other effectors, GRKs phosphorylate activated receptors, and arrestins subsequently bind phosphorylated receptors and cause receptor desensitization. Arrestins activated by interaction with phosphorylated receptors can also mediate G-protein-independent signalling by serving as adaptors to link receptors to numerous signalling pathways. Despite their central role in regulation and signalling of GPCRs, a structural understanding of β-arrestin activation and interaction with GPCRs is still lacking. Here we report the crystal structure of β-arrestin-1 (also called arrestin-2) in complex with a fully phosphorylated 29-amino-acid carboxy-terminal peptide derived from the human V2 vasopressin receptor (V2Rpp). This peptide has previously been shown to functionally and conformationally activate β-arrestin-1 (ref. 5). To capture this active conformation, we used a conformationally selective synthetic antibody fragment (Fab30) that recognizes the phosphopeptide-activated state of β-arrestin-1. The structure of the β-arrestin-1-V2Rpp-Fab30 complex shows marked conformational differences in β-arrestin-1 compared to its inactive conformation. These include rotation of the amino- and carboxy-terminal domains relative to each other, and a major reorientation of the 'lariat loop' implicated in maintaining the inactive state of β-arrestin-1. These results reveal, at high resolution, a receptor-interacting interface on β-arrestin, and they indicate a potentially general molecular mechanism for activation of these multifunctional signalling and regulatory proteins.
Figures
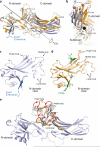
Figure 1. Fab30 specifically recognizes and stabilizes an active state of β-arrestin1
a, G protein coupled receptors are phosphorylated following activation, leading to the binding of arrestins. Interactions between the phosphorylated receptor and β-arrestin1 lead to β-arrestin1 activation and the subsequent blockade of G protein signaling and initiation of β-arrestin1 signaling pathways. b, Interaction between β-arrestin1 and Fab30 requires the presence of V2Rpp in a size exclusion assay. c, The formation of a complex between a GPCR and β-arrestin allosterically leads to an enhanced affinity of agonist for the receptor, termed the “high agonist affinity state.” Therefore, the fraction of receptor in the high agonist affinity state reflects the extent of complex formation between receptor and β-arrestin. In a radioligand competition binding assay using 125I-cyanopindolol as the probe and the agonist isoproterenol (Iso) as the competitor, β-arrestin1 alone shifts a small portion (14%) of receptors into the high agonist affinity state. Fab30 significantly amplifies this effect (31%) (n=3, p<0.0001 in F test). d, In a pull-down assay, phosphorylated β2-V2R chimera shows appreciable binding to β-arrestin1 only in the presence of Fab30. e, Overall structure of the β-arrestin1:V2Rpp:Fab30 complex.
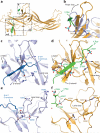
Figure 2. Conformational changes associated with β-arrestin1 activation
The structures of inactive β-arrestin1 (PDB ID: 1G4M chain A, light blue) and active β-arrestin1 (gold) were aligned on the N-domains. The β-arrestin1 carboxy terminus is highlighted in dark blue. a, A substantial rotation and translation of the C-domain relative to the N-domain occurs upon activation. The rotation axis is indicated as a solid black line. b, View of C-domain rotation along the axis. c, N-domain of inactive arrestin, highlighting important regions. d, Active β-arrestin1 in the same orientation, showing V2Rpp in green. Phosphorylated residues are highlighted as sticks. e, The overall structure of inactive β-arrestin1 (PDB ID: 1G4M, chain A), with loops from all inactive β-arrestin1 structures superimposed (grey loops). The active conformation of these loops (orange loops) deviates from all inactive structures.
Figure 3. V2Rpp interactions with β-arrestin1
a, Overall view of β-arrestin1, with regions of interest in boxes. Select charge-charge contacts are shown in dotted lines. b, V2Rpp (green) displaces the inactive finger loop (light blue), causing it to adopt an extended conformation in the active state (gold). c, In the inactive conformation, the β-arrestin1 carboxy-terminal β strand (dark blue) lies along the N-domain in the “three element” interaction network. d, Upon activation, this strand is displaced by the carboxy terminus of the V2Rpp, which engages in extensive charge-charge interactions through phosphorylated residues. e, The “polar core” of β-arrestin1 is thought to be a critical stabilizer of the inactive state. f, Upon V2Rpp binding, the carboxy-terminal strand residue Arg393 is displaced, and its interaction partner D297 undergoes a large movement together with the rest of the lariat loop.
Comment in
- Structural biology: Active arrestin proteins crystallized.
Borshchevskiy V, Büldt G. Borshchevskiy V, et al. Nature. 2013 May 2;497(7447):45-6. doi: 10.1038/nature12096. Epub 2013 Apr 24. Nature. 2013. PMID: 23604255 No abstract available. - Cell signalling: Crystallizing active arrestins.
Wrighton KH. Wrighton KH. Nat Rev Mol Cell Biol. 2013 Jun;14(6):326. doi: 10.1038/nrm3592. Epub 2013 May 15. Nat Rev Mol Cell Biol. 2013. PMID: 23673968 No abstract available.
Similar articles
- β-Arrestin biosensors reveal a rapid, receptor-dependent activation/deactivation cycle.
Nuber S, Zabel U, Lorenz K, Nuber A, Milligan G, Tobin AB, Lohse MJ, Hoffmann C. Nuber S, et al. Nature. 2016 Mar 31;531(7596):661-4. doi: 10.1038/nature17198. Epub 2016 Mar 23. Nature. 2016. PMID: 27007855 Free PMC article. - Crystal structure of pre-activated arrestin p44.
Kim YJ, Hofmann KP, Ernst OP, Scheerer P, Choe HW, Sommer ME. Kim YJ, et al. Nature. 2013 May 2;497(7447):142-6. doi: 10.1038/nature12133. Epub 2013 Apr 21. Nature. 2013. PMID: 23604253 - Activation-dependent conformational changes in {beta}-arrestin 2.
Xiao K, Shenoy SK, Nobles K, Lefkowitz RJ. Xiao K, et al. J Biol Chem. 2004 Dec 31;279(53):55744-53. doi: 10.1074/jbc.M409785200. Epub 2004 Oct 22. J Biol Chem. 2004. PMID: 15501822 - β-arrestins and G protein-coupled receptor trafficking.
Tian X, Kang DS, Benovic JL. Tian X, et al. Handb Exp Pharmacol. 2014;219:173-86. doi: 10.1007/978-3-642-41199-1_9. Handb Exp Pharmacol. 2014. PMID: 24292830 Free PMC article. Review. - Many faces of the GPCR-arrestin interaction.
Kim K, Chung KY. Kim K, et al. Arch Pharm Res. 2020 Sep;43(9):890-899. doi: 10.1007/s12272-020-01263-w. Epub 2020 Aug 14. Arch Pharm Res. 2020. PMID: 32803684 Review.
Cited by
- C-terminal threonines and serines play distinct roles in the desensitization of rhodopsin, a G protein-coupled receptor.
Azevedo AW, Doan T, Moaven H, Sokal I, Baameur F, Vishnivetskiy SA, Homan KT, Tesmer JJ, Gurevich VV, Chen J, Rieke F. Azevedo AW, et al. Elife. 2015 Apr 24;4:e05981. doi: 10.7554/eLife.05981. Elife. 2015. PMID: 25910054 Free PMC article. - Membrane phosphoinositides regulate GPCR-β-arrestin complex assembly and dynamics.
Janetzko J, Kise R, Barsi-Rhyne B, Siepe DH, Heydenreich FM, Kawakami K, Masureel M, Maeda S, Garcia KC, von Zastrow M, Inoue A, Kobilka BK. Janetzko J, et al. Cell. 2022 Nov 23;185(24):4560-4573.e19. doi: 10.1016/j.cell.2022.10.018. Epub 2022 Nov 10. Cell. 2022. PMID: 36368322 Free PMC article. - Location-biased β-arrestin conformations direct GPCR signaling.
Pham U, Chundi A, Stępniewski TM, Darbha S, Eiger DS, Gazula S, Gardner J, Hicks C, Selent J, Rajagopal S. Pham U, et al. bioRxiv [Preprint]. 2024 Sep 26:2024.09.24.614742. doi: 10.1101/2024.09.24.614742. bioRxiv. 2024. PMID: 39386521 Free PMC article. Preprint. - Crystal Structure of β-Arrestin 2 in Complex with CXCR7 Phosphopeptide.
Min K, Yoon HJ, Park JY, Baidya M, Dwivedi-Agnihotri H, Maharana J, Chaturvedi M, Chung KY, Shukla AK, Lee HH. Min K, et al. Structure. 2020 Sep 1;28(9):1014-1023.e4. doi: 10.1016/j.str.2020.06.002. Epub 2020 Jun 23. Structure. 2020. PMID: 32579945 Free PMC article. - GPCR-G Protein-β-Arrestin Super-Complex Mediates Sustained G Protein Signaling.
Thomsen ARB, Plouffe B, Cahill TJ 3rd, Shukla AK, Tarrasch JT, Dosey AM, Kahsai AW, Strachan RT, Pani B, Mahoney JP, Huang L, Breton B, Heydenreich FM, Sunahara RK, Skiniotis G, Bouvier M, Lefkowitz RJ. Thomsen ARB, et al. Cell. 2016 Aug 11;166(4):907-919. doi: 10.1016/j.cell.2016.07.004. Epub 2016 Aug 4. Cell. 2016. PMID: 27499021 Free PMC article.
References
- Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–650. doi:10.1038/nrm908. - PubMed
- Hepler JR, Gilman AG. G proteins. Trends Biochem Sci. 1992;17:383–387. - PubMed
- Freedman NJ, Lefkowitz RJ. Desensitization of G protein-coupled receptors. Recent Prog Horm Res. 1996;51:319–351. discussion 352-313. - PubMed
- Lefkowitz RJ, Shenoy SK. Transduction of receptor signals by beta-arrestins. Science. 2005;308:512–517. doi:10.1126/science.1109237. - PubMed
- Nobles KN, Guan Z, Xiao K, Oas TG, Lefkowitz RJ. The active conformation of beta-arrestin1: direct evidence for the phosphate sensor in the N-domain and conformational differences in the active states of beta-arrestins1 and -2. J Biol Chem. 2007;282:21370–21381. doi:10.1074/jbc.M611483200. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 GM074946/GM/NIGMS NIH HHS/United States
- HL 075443/HL/NHLBI NIH HHS/United States
- U01 GM094588/GM/NIGMS NIH HHS/United States
- P41 RR011823/RR/NCRR NIH HHS/United States
- R01 HL016037/HL/NHLBI NIH HHS/United States
- U54 GM087519/GM/NIGMS NIH HHS/United States
- GM087519/GM/NIGMS NIH HHS/United States
- P01 HL075443/HL/NHLBI NIH HHS/United States
- R01 HL070631/HL/NHLBI NIH HHS/United States
- HL70631/HL/NHLBI NIH HHS/United States
- R37 NS028471/NS/NINDS NIH HHS/United States
- HL16037/HL/NHLBI NIH HHS/United States
- R01 GM072688/GM/NIGMS NIH HHS/United States
- R01 NS028471/NS/NINDS NIH HHS/United States
- GM072688/GM/NIGMS NIH HHS/United States
- NS028471/NS/NINDS NIH HHS/United States
- HHMI_/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous