Heterogeneity of tumor-induced gene expression changes in the human metabolic network - PubMed (original) (raw)
Heterogeneity of tumor-induced gene expression changes in the human metabolic network
Jie Hu et al. Nat Biotechnol. 2013 Jun.
Abstract
Reprogramming of cellular metabolism is an emerging hallmark of neoplastic transformation. However, it is not known how the expression of metabolic genes in tumors differs from that in normal tissues, or whether different tumor types exhibit similar metabolic changes. Here we compare expression patterns of metabolic genes across 22 diverse types of human tumors. Overall, the metabolic gene expression program in tumors is similar to that in the corresponding normal tissues. Although expression changes of some metabolic pathways (e.g., upregulation of nucleotide biosynthesis and glycolysis) are frequently observed across tumors, expression changes of other pathways (e.g., oxidative phosphorylation) are very heterogeneous. Our analysis also suggests that the expression changes of some metabolic genes (e.g., isocitrate dehydrogenase and fumarate hydratase) may enhance or mimic the effects of recurrent mutations in tumors. On the level of individual biochemical reactions, many hundreds of metabolic isoenzymes show significant and tumor-specific expression changes. These isoenzymes are potential targets for anticancer therapy.
Figures
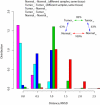
Figure 1
Global differences in metabolic gene expression between tumors and normal tissues. Colors represent distributions of the Euclidean (RMSD) expression distance between different samples of identical normal tissues (Normal_n_-Normal_n_, magenta), different samples of identical tumors (Tumor_n_-Tumor_n_, cyan), tumors and corresponding normal tissues (Tumor_n_-Normal_n_, blue), different tumors (Tumor_n_-Tumor_m_, green), and different normal tissues (Normal_n_-Normal_m_, red). The distributions shown in the figure were binned for display purposes only. Inset summarizes the average distances between pairs of tissues as a percentage of the average distance between two different normal tissues.
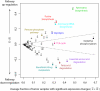
Figure 2
Expression of individual metabolic pathways in tumors. The biochemical pathways defined in the KEGG database (see Supplementary Table 5 for pathway numbering) are shown in the coordinates of (n¯+m¯, horizontal axis) and (n¯−m¯, vertical axis), where n¯ is the average fraction of tumor samples in which a pathway is significantly up-regulated, and m¯ is the average fraction in which a pathway is significantly down-regulated. The averages n¯ and m¯ were calculated across all 22 tumors. The up- (down-) regulation significance was determined using Wilcoxon signed-rank test (FDR-corrected P-value < 0.05, see Supplementary Fig. 4b for the same analysis with FDR = 0.2). Several pathways are highlighted using different colors. The dashed lines demarcate the region where n¯−m¯ is less than 20% of n¯+m¯ and are shown for visualization purposes only. Metabolic pathways without significant expression changes are primarily clustered on the left of the figure. Pathways that are often significantly up-regulated (high n¯ values) occupy positions in the upper right corner, while pathways that are primarily down-regulated (high m¯ values) occupy positions in the lower right corner. Highly heterogeneous pathways that show, in different tumors, both significant up- and down-regulation are clustered on the right near zero on the vertical axis.
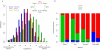
Figure 3
Tumor-induced mRNA expression changes for individual biochemical reactions in central metabolism. (a) Each metabolic reaction is marked with the number of tumors (out of 22 considered in our analysis) in which at least one isoenzyme catalyzing the corresponding reaction is significantly (FDR-corrected P-value < 0.05) up-regulated (red) and down-regulated (blue). (b) Reactions that are significantly up-regulated (red triangles) or down-regulated (blue triangles) when all isoenzymes and members of the corresponding protein complexes are considered together across all tumors (deep red or deep blue, FDR-corrected P-value < 0.05; light red or light blue, FDR-corrected P-value < 0.1). If unmarked, no statistically significant change in mRNA expression was detected.
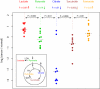
Figure 4
Cancer-induced changes in relative isoenzyme expression. (a) The Kullback-Leibler (KL) divergence was used to characterize differences in the relative expression of isoenzymes for all biochemical reactions with multiple isoenzymes. Colors represent distributions of the KL divergence in isoenzyme expression between different samples of identical normal tissues (Normal_n_-Normal_n_, blue), different samples of identical tumors (Tumor_n_-Tumor_n_, red), and tumors and corresponding normal tissues (Tumor_n_-Normal_n_, green). Inset summarizes the average KL divergences between pairs of tissues as a percentage of the average KL divergence between different samples of identical normal tissues. (b) Relative expression of the aldolase isoenzymes for kidney, liver, stomach, brain (GBM) tumors and the corresponding normal tissues.
Figure 5
Concentration changes for measured metabolites of the TCA cycle. The metabolite data, obtained from 10 colon cancer patients, contained matched normal and tumor samples. Every point in the figure represents the log2 ratio of tumor-to-normal concentration change for a single patient. The P-values above double arrows (in black) indicate the Wilcoxon signed-rank test significance of changes between consecutive metabolites. The P-values below metabolite names (in colors) indicate the Wilcoxon signed-rank test significance of changes between matched normal and tumor samples. The inset shows the measured metabolites in the context of the TCA cycle.
Comment in
- Cancer metabolism in breadth and depth.
Chun MG, Shaw RJ. Chun MG, et al. Nat Biotechnol. 2013 Jun;31(6):505-7. doi: 10.1038/nbt.2611. Nat Biotechnol. 2013. PMID: 23752435 No abstract available.
Similar articles
- Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer.
Sciacovelli M, Frezza C. Sciacovelli M, et al. FEBS J. 2017 Oct;284(19):3132-3144. doi: 10.1111/febs.14090. Epub 2017 May 21. FEBS J. 2017. PMID: 28444969 Free PMC article. Review. - Genetically-defined metabolic reprogramming in cancer.
Mullen AR, DeBerardinis RJ. Mullen AR, et al. Trends Endocrinol Metab. 2012 Nov;23(11):552-9. doi: 10.1016/j.tem.2012.06.009. Epub 2012 Jul 31. Trends Endocrinol Metab. 2012. PMID: 22858391 Free PMC article. Review. - The implications of IDH mutations for cancer development and therapy.
Pirozzi CJ, Yan H. Pirozzi CJ, et al. Nat Rev Clin Oncol. 2021 Oct;18(10):645-661. doi: 10.1038/s41571-021-00521-0. Epub 2021 Jun 15. Nat Rev Clin Oncol. 2021. PMID: 34131315 Review. - Metabolic alteration in tumorigenesis.
Yang H, Xiong Y, Guan K. Yang H, et al. Sci China Life Sci. 2013 Dec;56(12):1067-75. doi: 10.1007/s11427-013-4549-2. Epub 2013 Oct 10. Sci China Life Sci. 2013. PMID: 24114443 Review. - The phosphatidylinositol 3,5-bisphosphate (PI(3,5)P2)-dependent Tup1 conversion (PIPTC) regulates metabolic reprogramming from glycolysis to gluconeogenesis.
Han BK, Emr SD. Han BK, et al. J Biol Chem. 2013 Jul 12;288(28):20633-45. doi: 10.1074/jbc.M113.452813. Epub 2013 Jun 3. J Biol Chem. 2013. PMID: 23733183 Free PMC article.
Cited by
- The space of enzyme regulation in HeLa cells can be inferred from its intracellular metabolome.
Diener C, Muñoz-Gonzalez F, Encarnación S, Resendis-Antonio O. Diener C, et al. Sci Rep. 2016 Jun 23;6:28415. doi: 10.1038/srep28415. Sci Rep. 2016. PMID: 27335086 Free PMC article. - Candidate SNP Markers of Chronopathologies Are Predicted by a Significant Change in the Affinity of TATA-Binding Protein for Human Gene Promoters.
Ponomarenko P, Rasskazov D, Suslov V, Sharypova E, Savinkova L, Podkolodnaya O, Podkolodny NL, Tverdokhleb NN, Chadaeva I, Ponomarenko M, Kolchanov N. Ponomarenko P, et al. Biomed Res Int. 2016;2016:8642703. doi: 10.1155/2016/8642703. Epub 2016 Aug 22. Biomed Res Int. 2016. PMID: 27635400 Free PMC article. - AI hybrid survival assessment for advanced heart failure patients with renal dysfunction.
Zhang G, Wang Z, Tong Z, Qin Z, Su C, Li D, Xu S, Li K, Zhou Z, Xu Y, Zhang S, Wu R, Li T, Zheng Y, Zhang J, Cheng K, Tang J. Zhang G, et al. Nat Commun. 2024 Aug 8;15(1):6756. doi: 10.1038/s41467-024-50415-9. Nat Commun. 2024. PMID: 39117613 Free PMC article. - Data-Driven Metabolic Pathway Compositions Enhance Cancer Survival Prediction.
Auslander N, Wagner A, Oberhardt M, Ruppin E. Auslander N, et al. PLoS Comput Biol. 2016 Sep 27;12(9):e1005125. doi: 10.1371/journal.pcbi.1005125. eCollection 2016 Sep. PLoS Comput Biol. 2016. PMID: 27673682 Free PMC article. - Immunomodulatory effect of tibetan medicine compound extracts against ORFV in vitro by metabolomics.
Fan Y, Wu J, Huang W, Li S, Zeng Q, Gesang Z, Silang Y, Zhang C, Fu G. Fan Y, et al. BMC Vet Res. 2024 Aug 15;20(1):366. doi: 10.1186/s12917-024-04204-7. BMC Vet Res. 2024. PMID: 39143608 Free PMC article.
References
- Warburg O, Posener K, Negelein E. On the metabolism of carcinoma cells. Biochem Z. 1924;152:309–344.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. - PubMed
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM079759/GM/NIGMS NIH HHS/United States
- R00 CA168997/CA/NCI NIH HHS/United States
- R01ES019319/ES/NIEHS NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- GM079759/GM/NIGMS NIH HHS/United States
- R01 ES019319/ES/NIEHS NIH HHS/United States
- R00CA168997/CA/NCI NIH HHS/United States
- U54CA121852/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources