Monoclonal antibodies recognize gly-leu-phe-gly repeat of nucleoporin nup98 of tetrahymena, yeasts, and humans - PubMed (original) (raw)
Monoclonal antibodies recognize gly-leu-phe-gly repeat of nucleoporin nup98 of tetrahymena, yeasts, and humans
Masaaki Iwamoto et al. Monoclon Antib Immunodiagn Immunother. 2013 Apr.
Abstract
Nucleoporin Nup98, an essential component of the nuclear pore complex, has multifunctional roles in nuclear functions including transcriptional regulation and nucleocytoplasmic transport. These functions mostly depend on a Gly-Leu-Phe-Gly (GLFG) sequence appearing repetitively in the N-terminal region of Nup98. As the GLFG sequence is well conserved among Nup98s from a wide variety of species including humans, yeasts, and ciliates such as Tetrahymena thermophila, a specific antibody that recognizes the GLFG sequence is expected to detect various Nup98s from a wide-range of species. To generate monoclonal antibodies specific to the GLFG repeat of Nup98, we used two synthetic polypeptides derived from the macronuclear Nup98 of T. thermophila as an antigen. We obtained two monoclonal antibodies (MAbs), 13C2 and 21A10, that recognize Nup98s in indirect immunofluorescence staining and Western blot analysis of T. thermophila. Peptide array analysis of these monoclonal antibodies located the position of their epitopes at or near GLFG residues: the epitope recognized by the 13C2 MAb is FGxxN (x being any amino acid), and the epitope recognized by the 21A10 MAb is GLF. As expected by their epitopes, these monoclonal antibodies also recognize Nup98 homologs expressed by human cells and the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae, indicating that 13C2 and 21A10 MAbs recognize Nup98 epitopes common to phylogenetically distinct organisms. Thus, these MAbs are useful in studying a wide variety of biological phenomena that involve Nup98, ranging from ciliate nuclear dimorphism to NUP98-related human leukemia.
Figures
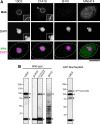
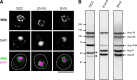
FIG. 1.
MAbs 13C2 and 21A10 detect MacNup98A of Tetrahymena. (A) IF staining of T. thermophila cells using MAbs 13C2, 21A10, 2H10, and 414. Black-and-white images were obtained with DAPI. Color images represent MAb (green) merged with DAPI (magenta). Dotted lines represent the outline of cells. Open arrow indicates the micronucleus. Insets are magnified images showing the position of the micronucleus. Bar, 20 μm. (B) WB analysis of T. thermophila specimen from wild-type (left lanes) or GFP-MacNup98A expressing cells (far right lane). MAbs used are indicated above each lane. Signals were detected using the same staining conditions, except that the exposure time of the middle lane (21A10 long exposure) was about 10 times longer than that of the other lanes. Open arrowheads and closed arrow show the positions of MacNup98A and GFP-MacNup98A, respectively. Diamonds and asterisks identify uncharacterized proteins.
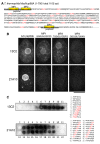
FIG. 2.
Determination of epitopes for 13C2 and 21A10 MAbs. (A) Amino acid sequence of MacNup98A from residue 1–700; the total length of MacNup98A is 1105 amino acids. The two regions (peptides 1 and 2) underlined are the sequences used for immunization of BALB/c mice. The sequences highlighted in yellow, designated MP1, MP2, and MP3, were synthesized for the antibody-masking experiments shown in B. Each GLFG is shown in red. (B) The epitope masking effect of none (w/o peptide), MP1 (GGG_GLFG_NTQ), MP2 (TGG_GLFG_QPQ), and MP3 (QGG_GLFG_AAN) was examined by IF staining using MAbs 13C2 (upper panel) and 21A10 (lower panel). MAbs were incubated with one of the oligopeptides for 30 min prior to incubation with fixed T. thermophila cells. Bar, 20 μm. (C) Peptide array analysis using MAbs 13C2 (top) or 21A10 (bottom). Numbers at the top and bottom of the membrane correspond to those listed on the right; each of these 10-mer peptides was tested for binding to MAbs 13C2 and 21A10, as described in Methods section. Residues shown in red are common sequences among the peptides detected by each MAb.
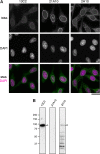
FIG. 3.
MAbs 13C2 and 21A10 cross-react with Nup98 of HeLa cell. (A) IF staining of HeLa cells using MAbs 13C2, 21A10, or 2H10. Black-and-white images represent fluorescence images obtained with MAb (top) and DAPI (middle). Color images represent MAb (green) merged with DAPI (magenta). Bar, 20 μm. (B) WB analysis of HeLa cells using 13C2, 21A10, or 2H10. The blots were treated identically; the exposure time for chemiluminescence detection was 10 min. Open arrows represent the position of Nup98. The lower bands appearing in the 2H10 lane were non-specific staining with the anti-rat IgG secondary antibody (data not shown).
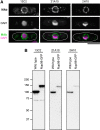
FIG. 4.
MAbs 13C2 and 21A10 cross-react with Nup98 of fission yeast. (A) IF staining of S. pombe cells using MAbs 13C2, 21A10, and 2H10; 1:10 dilution of the supernatants of hybridoma cultures of 13C2 or 21A10 or 10 μg/mL IgG solution of 2H10 were used. Black-and-white images represent fluorescence images obtained with MAb (top) and DAPI (middle). Color images represent merged images of MAb (green) with DAPI (magenta). Dotted lines represent the outline of cells. Bar, 5 μm. (B) WB analysis of S. pombe cell extract using MAbs 13C2 (left lanes), 21A10 (middle lanes), and 2H10 (right lanes); 1:10 dilutions of the supernatants of hybridoma culture medium of 13C2 or 21A10 or 1 μg/mL IgG solution of 2H10 were used. Left and right lanes represent specimens from a wild-type strain and an S. pombe strain in which Nup98 was chromosomally replaced with Nup98-GFP,(31) respectively.
FIG. 5.
MAbs 13C2 and 21A10 crossreact with multiple nucleoporins of budding yeast. (A) IF staining of S. cereviciae cells using MAbs 13C2, 21A10, and 2H10; 1:10 dilution of the supernatants of hybridoma cultures of 13C2 or 21A10 or 10 μg/mL IgG solution of 2H10 were used. Black-and-white images represent fluorescence results obtained with MAb (top) and DAPI (middle). Color images represent MAb (green) merged with DAPI (magenta). Dotted lines represent the outline of cells. Bar, 5 μm. (B) WB analysis of S. cereviciae cell extract using MAbs 13C2, 21A10, or 2H10; 1:10 dilution of the supernatants of hybridoma culture medium of 13C2 or 21A10 or 2 μg/mL IgG solution of 2H10 were used. Arrows represent the positions of the indicated nucleoporins. Asterisks represent uncharacterized proteins.
Similar articles
- Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena.
Iwamoto M, Mori C, Kojidani T, Bunai F, Hori T, Fukagawa T, Hiraoka Y, Haraguchi T. Iwamoto M, et al. Curr Biol. 2009 May 26;19(10):843-7. doi: 10.1016/j.cub.2009.03.055. Epub 2009 Apr 16. Curr Biol. 2009. PMID: 19375312 - Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms.
Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Iwamoto M, et al. Genes Cells. 2010 Jun;15(7):661-9. doi: 10.1111/j.1365-2443.2010.01415.x. Epub 2010 Jun 7. Genes Cells. 2010. PMID: 20545767 Review. - Formation of Nup98-containing nuclear bodies in HeLa sublines is linked to genomic rearrangements affecting chromosome 11.
Romana S, Radford-Weiss I, Lapierre JM, Doye V, Geoffroy MC. Romana S, et al. Chromosoma. 2016 Sep;125(4):789-805. doi: 10.1007/s00412-015-0567-0. Epub 2015 Dec 21. Chromosoma. 2016. PMID: 26685999 - Domain topology of nucleoporin Nup98 within the nuclear pore complex.
Chatel G, Desai SH, Mattheyses AL, Powers MA, Fahrenkrog B. Chatel G, et al. J Struct Biol. 2012 Jan;177(1):81-9. doi: 10.1016/j.jsb.2011.11.004. Epub 2011 Nov 12. J Struct Biol. 2012. PMID: 22100335 Free PMC article. - NUP98 fusion in human leukemia: dysregulation of the nuclear pore and homeodomain proteins.
Nakamura T. Nakamura T. Int J Hematol. 2005 Jul;82(1):21-7. doi: 10.1532/IJH97.04160. Int J Hematol. 2005. PMID: 16105755 Review.
Cited by
- Asymmetrical localization of Nup107-160 subcomplex components within the nuclear pore complex in fission yeast.
Asakawa H, Kojidani T, Yang HJ, Ohtsuki C, Osakada H, Matsuda A, Iwamoto M, Chikashige Y, Nagao K, Obuse C, Hiraoka Y, Haraguchi T. Asakawa H, et al. PLoS Genet. 2019 Jun 6;15(6):e1008061. doi: 10.1371/journal.pgen.1008061. eCollection 2019 Jun. PLoS Genet. 2019. PMID: 31170156 Free PMC article. - Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in the ciliate Tetrahymena.
Iwamoto M, Osakada H, Mori C, Fukuda Y, Nagao K, Obuse C, Hiraoka Y, Haraguchi T. Iwamoto M, et al. J Cell Sci. 2017 May 15;130(10):1822-1834. doi: 10.1242/jcs.199398. Epub 2017 Apr 6. J Cell Sci. 2017. PMID: 28386019 Free PMC article. - Uncleavable Nup98-Nup96 is functional in the fission yeast Schizosaccharomyces pombe.
Asakawa H, Mori C, Ohtsuki C, Iwamoto M, Hiraoka Y, Haraguchi T. Asakawa H, et al. FEBS Open Bio. 2015 Jun 12;5:508-14. doi: 10.1016/j.fob.2015.06.004. eCollection 2015. FEBS Open Bio. 2015. PMID: 26137436 Free PMC article. - Novel Therapeutic Targets in Acute Myeloid Leukemia (AML).
Wysota M, Konopleva M, Mitchell S. Wysota M, et al. Curr Oncol Rep. 2024 Apr;26(4):409-420. doi: 10.1007/s11912-024-01503-y. Epub 2024 Mar 19. Curr Oncol Rep. 2024. PMID: 38502417 Free PMC article. Review. - Characterization of nuclear pore complex components in fission yeast Schizosaccharomyces pombe.
Asakawa H, Yang HJ, Yamamoto TG, Ohtsuki C, Chikashige Y, Sakata-Sogawa K, Tokunaga M, Iwamoto M, Hiraoka Y, Haraguchi T. Asakawa H, et al. Nucleus. 2014 Mar-Apr;5(2):149-62. doi: 10.4161/nucl.28487. Epub 2014 Mar 12. Nucleus. 2014. PMID: 24637836 Free PMC article.
References
- Yang Q. Rout MP. Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. - PubMed
- Hoelz A. Debler EW. Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials