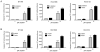The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis - PubMed (original) (raw)
The major reverse transcriptase-incompetent splice variant of the human telomerase protein inhibits telomerase activity but protects from apoptosis
Imke Listerman et al. Cancer Res. 2013.
Abstract
Human telomerase reverse transcriptase (hTERT; the catalytic protein subunit of telomerase) is subjected to numerous alternative splicing events, but the regulation and function of these splice variants is obscure. Full-length hTERT includes conserved domains that encode reverse transcriptase activity, RNA binding, and other functions. The major splice variant termed α+β- or β-deletion is highly expressed in stem and cancer cells, where it codes for a truncated protein lacking most of the reverse transcriptase domain but retaining the known RNA-binding motifs. In a breast cancer cell panel, we found that β-deletion was the hTERT transcript that was most highly expressed. Splicing of this transcript was controlled by the splice regulators SRSF11, HNRNPH2, and HNRNPL, and the β-deletion transcript variant was associated with polyribosomes in cells. When ectopically overexpressed, β-deletion protein competed for binding to telomerase RNA (hTR/TERC), thereby inhibiting endogenous telomerase activity. Overexpressed β-deletion protein localized to the nucleus and mitochondria and protected breast cancer cells from cisplatin-induced apoptosis. Our results reveal that a major hTERT splice variant can confer a growth advantage to cancer cells independent of telomere maintenance, suggesting that hTERT makes multiple contributions to cancer pathophysiology.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures
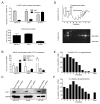
Figure 1. Schematic of hTERT α/βsplice variants
Top: hTERT protein domain structure. Below: Full-length hTERT mRNA, α/β splice variants, drawn as open boxes and approximately to scale. Black arrow indicates open reading frame. Picture adapted from (6).
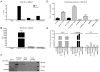
Figure 2. The β-deletion splice variant can escape NMD and is associated with polysomes
A, Top: hTERT α+β+ and β-deletion mRNA splice variant expression in copies/μg RNA, normalized to copies/μg RNA GAPDH mRNA in UM-UC-3, Jurkat and BT-549 cells. Below: Relative telomerase activity (RTA) in 2500 cells/μl. B, Accumulation of hTERT splice variant and SRSF3 PTC+ mRNAs upon UPF1 versus control shRNA knockdown. Error bars are SD from at least 3 experiments. C, Western Blot of UPF1 and GAPDH in cells treated with UPF1 or control shRNAs. D, Top: Representative absorbance profile for RNA separated by velocity sedimentation through a 10–50% sucrose gradient. Positions of 40S, 60S, 80S, and polysomal peaks are indicated. Below: Agarose gel electrophoresis of RNA extracted from each fraction. 28S and 18S rRNAs are indicated. E, Abundance of SRSF3 and hTERT variant mRNA were measured by RT-qPCR and visualized as % SRSF3 PTC+/total SRSF3 and % β-deletion/α+containing hTERT mRNA.
Figure 3. The β-deletion isoform localizes to the nucleus, nucleuolus and mitochondria
β-deletion FLAG/GFP constructs were transfected into HeLa cells and stained with Hoechst 33342 and mitotracker deep red (Molecular Probes).
Figure 4. SRSF11, hnRNPH2 and hnRNPL regulate hTERT β-deletion splicing
A, Top: Structure of the pSpliceExpress-hTERT reporter gene. SRSF11 and hnRNPH2 binding sites are indicated with asterisks or plus signs, respectively. Below: qPCR primers for α+β+/β-deletion hTERT variants. RT primer for reporter gene-derived hTERT mRNA anneals to rat insulin exon 3. B, qRT-PCR of hTERT variants from RNA extracted from HEK293T cells co-transfected with either empty plasmid, splicing factor proteins and pSpliceExpress-hTERT. Bars represent mean of % β site exclusion. Error bars are SEM from 3 biological replicates assayed in triplicate. C, Western blot of HEK293T cell extracts extracts confirms overexpression of splicing factors.
Figure 5. The β-deletion isoform is a dominant-negative inhibitor of telomerase by sequestering hTR
A, hTR RNA associated with FLAG-hTERT constructs in GM847 cells. RNA from a parallel anti-FLAG immunoprecipitates from B was analyzed for presence of hTR and GAPDH RNA by RT-qPCR and repesented as % recovered over input RNA. B, RTA associated with anti-FLAG immunoprecipitates from GM847 cells transduced with indicated lentivirual constructs. C, Western blot of GM847 telomerase RNP immunoprecipitates used in Figure 5B. D, RTA in UM-UC-3 bladder cancer cells transduced with indicated lentiviral vectors. E, RNA from a parallel sample from D was extracted and analyzed for hTR and hTERT variant RNA normalized to GAPDH and vector control. Error bars in B and D represent SD from at least 3 experiments.
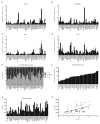
Figure 6. Expression of α/β splice variants, telomerase activity and telomere length in 50 breast cancer cell lines
A–D, Levels of indicated splice variants, expressed as transcript numbers normalized to GAPDH transcript numbers x 104. E, Relative expression of individual α/β hTERT splice variants relative to total amount of hTERT transcripts. F, RTA was measured by RQ-TRAP. G, Modal telomere length was determined by Southern blot from telomere restriction fragments (TRF). H, Regression of log(RTA) and TRF reveals a linear relationship (r = 0.487, P < 0.0001). Results in A–D, F and G are the average of 2–4 biological replicates and assayed in triplicate reactions (A–D, F); error bars represent SD.
Figure 7. The β-deletion protein protects basal breast cancer cells from apoptosis
BT-549, HCC1806 and HCC3153 that stably overexpressed β-deletion or vector control for less than 2 weeks were treated with the indicated cisplatin concentration for 48 h. Bars represent averaged luciferase activity of caspase 3/7 reporter over cell confluency of 4 replicates; error bars represent SEM.
Similar articles
- Quantification of hTERT splice variants in melanoma by SYBR green real-time polymerase chain reaction indicates a negative regulatory role for the beta deletion variant.
Lincz LF, Mudge LM, Scorgie FE, Sakoff JA, Hamilton CS, Seldon M. Lincz LF, et al. Neoplasia. 2008 Oct;10(10):1131-7. doi: 10.1593/neo.08644. Neoplasia. 2008. PMID: 18813352 Free PMC article. - A Common Cancer Risk-Associated Allele in the hTERT Locus Encodes a Dominant Negative Inhibitor of Telomerase.
Killedar A, Stutz MD, Sobinoff AP, Tomlinson CG, Bryan TM, Beesley J, Chenevix-Trench G, Reddel RR, Pickett HA. Killedar A, et al. PLoS Genet. 2015 Jun 8;11(6):e1005286. doi: 10.1371/journal.pgen.1005286. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26053551 Free PMC article. - Changes of telomerase activity by alternative splicing of full-length and beta variants of hTERT in breast cancer patients.
Rha SY, Jeung HC, Park KH, Kim JJ, Chung HC. Rha SY, et al. Oncol Res. 2009;18(5-6):213-20. doi: 10.3727/096504009x12596189659123. Oncol Res. 2009. PMID: 20225759 - Physiological and pathological significance of human telomerase reverse transcriptase splice variants.
Bollmann FM. Bollmann FM. Biochimie. 2013 Nov;95(11):1965-70. doi: 10.1016/j.biochi.2013.07.031. Epub 2013 Aug 9. Biochimie. 2013. PMID: 23933091 Review. - Human telomerase reverse transcriptase regulation by DNA methylation, transcription factor binding and alternative splicing (Review).
Avin BA, Umbricht CB, Zeiger MA. Avin BA, et al. Int J Oncol. 2016 Dec;49(6):2199-2205. doi: 10.3892/ijo.2016.3743. Epub 2016 Oct 20. Int J Oncol. 2016. PMID: 27779655 Free PMC article. Review.
Cited by
- Role of telomere length in human carcinogenesis (Review).
Tsatsakis A, Oikonomopoulou T, Nikolouzakis TK, Vakonaki E, Tzatzarakis M, Flamourakis M, Renieri E, Fragkiadaki P, Iliaki E, Bachlitzanaki M, Karzi V, Katsikantami I, Kakridonis F, Hatzidaki E, Tolia M, Svistunov AA, Spandidos DA, Nikitovic D, Tsiaoussis J, Berdiaki A. Tsatsakis A, et al. Int J Oncol. 2023 Jul;63(1):78. doi: 10.3892/ijo.2023.5526. Epub 2023 May 26. Int J Oncol. 2023. PMID: 37232367 Free PMC article. Review. - Evolution of TERT-interacting lncRNAs: expanding the regulatory landscape of telomerase.
Nelson AD, Shippen DE. Nelson AD, et al. Front Genet. 2015 Sep 10;6:277. doi: 10.3389/fgene.2015.00277. eCollection 2015. Front Genet. 2015. PMID: 26442096 Free PMC article. Review. - TERT-Regulation and Roles in Cancer Formation.
Dratwa M, Wysoczańska B, Łacina P, Kubik T, Bogunia-Kubik K. Dratwa M, et al. Front Immunol. 2020 Nov 19;11:589929. doi: 10.3389/fimmu.2020.589929. eCollection 2020. Front Immunol. 2020. PMID: 33329574 Free PMC article. Review. - Insights into Telomerase/hTERT Alternative Splicing Regulation Using Bioinformatics and Network Analysis in Cancer.
Ludlow AT, Slusher AL, Sayed ME. Ludlow AT, et al. Cancers (Basel). 2019 May 14;11(5):666. doi: 10.3390/cancers11050666. Cancers (Basel). 2019. PMID: 31091669 Free PMC article. Review. - An antiapoptotic role for telomerase RNA in human immune cells independent of telomere integrity or telomerase enzymatic activity.
Gazzaniga FS, Blackburn EH. Gazzaniga FS, et al. Blood. 2014 Dec 11;124(25):3675-84. doi: 10.1182/blood-2014-06-582254. Epub 2014 Oct 15. Blood. 2014. PMID: 25320237 Free PMC article.
References
- Blackburn EH. Telomere states and cell fates. Nature. 2000;408:53–6. - PubMed
- Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–60. - PubMed
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, et al. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–5. - PubMed
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–95. - PubMed
- Sykorova E, Fajkus J. Structure-function relationships in telomerase genes. Biol Cell. 2009;101:375–92. 1 p following 92. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical