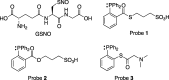Mechanism-based triarylphosphine-ester probes for capture of endogenous RSNOs - PubMed (original) (raw)
. 2013 May 22;135(20):7693-704.
doi: 10.1021/ja401565w. Epub 2013 May 8.
Affiliations
- PMID: 23614769
- PMCID: PMC3663071
- DOI: 10.1021/ja401565w
Mechanism-based triarylphosphine-ester probes for capture of endogenous RSNOs
Uthpala Seneviratne et al. J Am Chem Soc. 2013.
Abstract
Nitrosothiols (RSNOs) have been proposed as important intermediates in nitric oxide (NO(•)) metabolism, storage, and transport as well as mediators in numerous NO-signaling pathways. RSNO levels are finely regulated, and dysregulation is associated with the etiology of several pathologies. Current methods for RSNO quantification depend on indirect assays that limit their overall specificity and reliability. Recent developments of phosphine-based chemical probes constitute a promising approach for the direct detection of RSNOs. We report here results from a detailed mechanistic and kinetic study for trapping RSNOs by three distinct phosphine probes, including structural identification of novel intermediates and stability studies under physiological conditions. We further show that a triarylphosphine-thiophenyl ester can be used in the absolute quantification of endogenous GSNO in several cancer cell lines, while retaining the elements of the SNO functional group, using an LC-MS-based assay. Finally, we demonstrate that a common product ion (m/z = 309.0), derived from phosphine-RSNO adducts, can be used for the detection of other low-molecular weight nitrosothiols (LMW-RSNOs) in biological samples. Collectively, these findings establish a platform for the phosphine ligation-based, specific and direct detection of RSNOs in biological samples, a powerful tool for expanding the knowledge of the biology and chemistry of NO(•)-mediated phenomena.
Figures
Figure 1
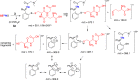
Structures of _S_-nitrosoglutathione (GSNO) and triarylphosphine probes used in this study.
Figure 2
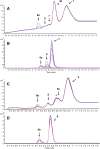
Reaction between GSNO and phosphine probe 1 (A, B) and 2 (C, D) (in excess, in Tris-HCl, pH 7.4), analyzed by LC–TOF-MS. Upper panel (A and C): time-dependent LC of the reaction intermediates and products (at 254 nm); bottom panel (B and D): time-dependent high-resolution ESI+-MS (EIC; extracted ion chromatogram). Colors: 23 min = black, 45 min = blue, 67 min = green, 89 min = purple, 111 min = yellow, 133 min = pink. Peaks corresponding to 1, 2, 4a, and 4b were excluded on LC–MS trace for clarity. GSNO, 5 μM and 50 μM with phosphine probe (20-fold excess) were used for the LC–MS and LC–UV studies, respectively, (*) indicates the hydrolyzed 2 under our experimental conditions. Monitored/expected masses, 5; m/z = 627.1678/627.1673 [M + H]+, 6a; m/z = 750.1376/750.1373 [M + H]+, 6b; m/z = 734.1602/734.1607 [M + H]+, 7; m/z = 322.0995/322.0991 [M + H]+, 8; m/z = 444.1029/444.1029 [M + H]+. _Y_-axis represents the relative intensity.
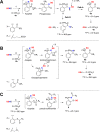
Scheme 1. Reaction Pathways Supported by 31P NMR and High-Resolution LC–MS Analysis of Intermediates and Products When GSNO Reacts with (A) Probe 1, (B) Probe 2, and (C) Probe 3 in Buffer Conditions at Physiological pH
Figure 3
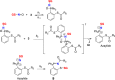
Capturing GSNO as disulfide-iminophosphorane (9) by probe 3. Time- and concentration-dependent TOF-MS analysis of the reaction mixture comprising GSNO (5 μM) with probe 3 (50 μM to 1 mM) in phosphate buffer (pH 5.7) at 24 °C. Colors: 2 min = black, 12 min = red, 22 min = blue, 32 min = green. When the concentration of probe 3 (1 mM) is in 200-fold molar excess, the formation of 9 occurs within 15 min with <10% of the phosphoryl-disulfide 10. Peaks corresponding to 3 and 4c were excluded on EIC for clarity. Monitored/expected masses, GSNO: m/z = 337.0810/337.0812/[M + H]+; 9: m/z = 350.6044/350.6048 [M + 2H]2+; 10: m/z = 616.1334/616.1336 [M + H]+. Phosphate buffer at pH 5.7 was used to increase ionization and to aid in detection of GSNO. _Y_-axis represents the relative intensity.
Scheme 2. 15N Fragments Derived from the Reaction of Probe 3 and GS15NO upon Collision Induced Dissociation (CID) in MS and Possible Resonance Stabilization of m/z = 309.0
Figure 4
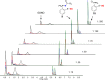
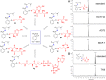
Disulfide-iminophosphorane 9 is formed in activated macrophage lysate (106 cells) upon treatment with probe 3. (A) LC–MS (TIC; total ion chromatogram) of cell lysate. (B) LC–MS/MS (MRM) of DTT-treated cell lysate (negative control). (C) MRM of cell lysate without treatment with probe 3, analyte corresponds to GSNO. (D) m/z = 350.6 → 309.0, (G) m/z = 350.6 → 487.1, (H) m/z = 350.6 → 571.1, and (E) internal standard (m/z = 352.1 → 309.0) in cell extracts. (F) Product ion spectra of 9, m/z = 350.6 [M + 2H]2+, derived from cell lysate (top), authentic standard (middle), and 15N fragment, m/z = 351.1 [M + 2H]2+, derived from the reaction of probe 3 and GS15NO (bottom). _Y_-axis represents the relative intensity.
Scheme 3. Proposed Kinetic Model for Capturing GSNO by Probe 3
Figure 5
(Left) Low-molecular weight nitrosothiols (LMW-RSNOs) that produce common product ion, m/z = 309.0, upon CID. (Right) Capturing LMW-RSNOs, by LC–MS/MS using the common product ion m/z 309.0 in cell lysates: (A) _S_-nitrosocysteine (9a) m/z = 309.0 → 257.6. (B) _S-_nitrosated _N_-acetyl-penicillamine (9h) m/z = 309.0 → 292.6. _Y_-axis represents the relative intensity.
Similar articles
- Chemical methods for the direct detection and labeling of S-nitrosothiols.
Bechtold E, King SB. Bechtold E, et al. Antioxid Redox Signal. 2012 Oct 1;17(7):981-91. doi: 10.1089/ars.2012.4570. Epub 2012 Mar 23. Antioxid Redox Signal. 2012. PMID: 22356122 Free PMC article. Review. - Water-soluble triarylphosphines as biomarkers for protein S-nitrosation.
Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, King SB. Bechtold E, et al. ACS Chem Biol. 2010 Apr 16;5(4):405-14. doi: 10.1021/cb900302u. ACS Chem Biol. 2010. PMID: 20146502 Free PMC article. - Understanding the Performance of Metal-Organic Frameworks for Modulation of Nitric Oxide Release from S-Nitrosothiols.
Ling P, Gao X, Zang X, Sun X, Gao F. Ling P, et al. Chem Asian J. 2022 Apr 1;17(7):e202101358. doi: 10.1002/asia.202101358. Epub 2022 Mar 3. Chem Asian J. 2022. PMID: 35178879 - Automated Online Solid-Phase Derivatization for Sensitive Quantification of Endogenous S-Nitrosoglutathione and Rapid Capture of Other Low-Molecular-Mass S-Nitrosothiols.
Wang X, Garcia CT, Gong G, Wishnok JS, Tannenbaum SR. Wang X, et al. Anal Chem. 2018 Feb 6;90(3):1967-1975. doi: 10.1021/acs.analchem.7b04049. Epub 2018 Jan 9. Anal Chem. 2018. PMID: 29271637 Free PMC article. - Novel method for measuring S-nitrosothiols using hydrogen sulfide.
Teng X, Scott Isbell T, Crawford JH, Bosworth CA, Giles GI, Koenitzer JR, Lancaster JR, Doeller JE, W Kraus D, P Patel R. Teng X, et al. Methods Enzymol. 2008;441:161-72. doi: 10.1016/S0076-6879(08)01209-3. Methods Enzymol. 2008. PMID: 18554534 Review.
Cited by
- The enzymatic function of the honorary enzyme: S-nitrosylation of hemoglobin in physiology and medicine.
Premont RT, Singel DJ, Stamler JS. Premont RT, et al. Mol Aspects Med. 2022 Apr;84:101056. doi: 10.1016/j.mam.2021.101056. Epub 2021 Nov 28. Mol Aspects Med. 2022. PMID: 34852941 Free PMC article. Review. - Mass spectrometry in studies of protein thiol chemistry and signaling: opportunities and caveats.
Baez NO, Reisz JA, Furdui CM. Baez NO, et al. Free Radic Biol Med. 2015 Mar;80:191-211. doi: 10.1016/j.freeradbiomed.2014.09.016. Epub 2014 Sep 28. Free Radic Biol Med. 2015. PMID: 25261734 Free PMC article. Review. - Conversion of S-phenylsulfonylcysteine residues to mixed disulfides at pH 4.0: utility in protein thiol blocking and in protein-S-nitrosothiol detection.
Reeves BD, Joshi N, Campanello GC, Hilmer JK, Chetia L, Vance JA, Reinschmidt JN, Miller CG, Giedroc DP, Dratz EA, Singel DJ, Grieco PA. Reeves BD, et al. Org Biomol Chem. 2014 Oct 28;12(40):7942-56. doi: 10.1039/c4ob00995a. Epub 2014 Jul 2. Org Biomol Chem. 2014. PMID: 24986430 Free PMC article. - Bioorthogonal Reactions of Triarylphosphines and Related Analogues.
Heiss TK, Dorn RS, Prescher JA. Heiss TK, et al. Chem Rev. 2021 Jun 23;121(12):6802-6849. doi: 10.1021/acs.chemrev.1c00014. Epub 2021 Jun 8. Chem Rev. 2021. PMID: 34101453 Free PMC article. Review. - Chemoproteomic Strategy to Quantitatively Monitor Transnitrosation Uncovers Functionally Relevant S-Nitrosation Sites on Cathepsin D and HADH2.
Zhou Y, Wynia-Smith SL, Couvertier SM, Kalous KS, Marletta MA, Smith BC, Weerapana E. Zhou Y, et al. Cell Chem Biol. 2016 Jun 23;23(6):727-37. doi: 10.1016/j.chembiol.2016.05.008. Epub 2016 Jun 9. Cell Chem Biol. 2016. PMID: 27291402 Free PMC article.
References
- Dedon P. C.; Tannenbaum S. R. Arch. Biochem. Biophys. 2004, 423, 12–22. - PubMed
- Keshive M.; Singh S.; Wishnok J. S.; Tannenbaum S. R.; Deen W. M. Chem. Res. Toxicol. 1996, 9, 988–993. - PubMed
- Dickinson D. A.; Forman H. J. Biochem. Pharmacol. 2002, 64, 1019–1026. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources