TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions - PubMed (original) (raw)
TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions
Daehwan Kim et al. Genome Biol. 2013.
Abstract
TopHat is a popular spliced aligner for RNA-sequence (RNA-seq) experiments. In this paper, we describe TopHat2, which incorporates many significant enhancements to TopHat. TopHat2 can align reads of various lengths produced by the latest sequencing technologies, while allowing for variable-length indels with respect to the reference genome. In addition to de novo spliced alignment, TopHat2 can align reads across fusion breaks, which can occur after genomic translocations. TopHat2 combines the ability to identify novel splice sites with direct mapping to known transcripts, producing sensitive and accurate alignments, even for highly repetitive genomes or in the presence of pseudogenes. TopHat2 is available at http://ccb.jhu.edu/software/tophat.
Figures
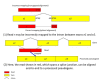
Figure 1
Two possible incorrect alignments of spliced reads. 1) A read extending a few bases into the flanking exon can be aligned to the intron instead of the exon. 2) A read spanning multiple exons from genes with processed pseudogene copies can be aligned to the pseudogene copies instead of the gene from which it originates.
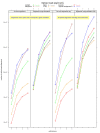
Figure 2
The number of read alignments from TopHat2, GSNAP, RUM, MapSplice, and STAR. Tthe RNA-seq reads are from Chen et al. [11]. TopHat2 was run with and without realignment (realignment edit distance of 0). TopHat2, GSNAP, and STAR were run in both de novo and gene-mapping modes, while MapSplice was run only in de novo mode and RUM was run only in gene-mapping mode. The number of alignments at each edit distance is cumulative; for instance, the number of alignments at an edit distance of 2 includes all the alignments with edit distance of 0, 1, or 2.
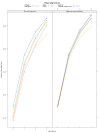
Figure 3
The number of spliced-read alignments from TopHat2, GSNAP, RUM, MapSplice, and STAR. The RNA-seq reads are from Chen et al. [11]. TopHat2, GSNAP, and STAR were run in both de novo and gene-mapping modes while MapSplice was run only in de novo mode and RUM was run only in gene-mapping mode. For each mapping mode, the two panels on the left show the number of spliced alignments whose splice sites were found in the gene annotations, and the two panels on the right show the number of all spliced alignments including novel splice sites.
Figure 4
The number of read and spliced-read alignments from TopHat2, using different realignment edit distances and no realignment. Edit distances of 0, 1, and 2 were used. As TopHat2 allows more realignment from no realignment to 2 to 1 to 0, the number of read alignments and spliced-read alignments increases, so that the differences in the numbers of read alignments from TopHat run with different realignment edit distance are mostly explained by the increase in the number of spliced-read alignments.
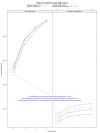
Figure 5
The number of spliced-read alignments from TopHat2, GSNAP, STAR, and MapSplice without using gene annotation. The number of read alignments whose splice sites were found in the gene annotations are shown in brown, and the number of all spliced-read alignments including novel splice sites are shown in green.
Figure 6
TopHat2 pipeline. Details are given in the main text.
Similar articles
- GeneScissors: a comprehensive approach to detecting and correcting spurious transcriptome inference owing to RNA-seq reads misalignment.
Zhang Z, Huang S, Wang J, Zhang X, Pardo Manuel de Villena F, McMillan L, Wang W. Zhang Z, et al. Bioinformatics. 2013 Jul 1;29(13):i291-9. doi: 10.1093/bioinformatics/btt216. Bioinformatics. 2013. PMID: 23812996 Free PMC article. - RNA-Seq read alignments with PALMapper.
Jean G, Kahles A, Sreedharan VT, De Bona F, Rätsch G. Jean G, et al. Curr Protoc Bioinformatics. 2010 Dec;Chapter 11:Unit 11.6. doi: 10.1002/0471250953.bi1106s32. Curr Protoc Bioinformatics. 2010. PMID: 21154708 - RNASequel: accurate and repeat tolerant realignment of RNA-seq reads.
Wilson GW, Stein LD. Wilson GW, et al. Nucleic Acids Res. 2015 Oct 15;43(18):e122. doi: 10.1093/nar/gkv594. Epub 2015 Jun 16. Nucleic Acids Res. 2015. PMID: 26082497 Free PMC article. - Mapping RNA-seq Reads with STAR.
Dobin A, Gingeras TR. Dobin A, et al. Curr Protoc Bioinformatics. 2015 Sep 3;51:11.14.1-11.14.19. doi: 10.1002/0471250953.bi1114s51. Curr Protoc Bioinformatics. 2015. PMID: 26334920 Free PMC article. Review. - Mapping RNA-seq reads to transcriptomes efficiently based on learning to hash method.
Yu X, Liu X. Yu X, et al. Comput Biol Med. 2020 Jan;116:103539. doi: 10.1016/j.compbiomed.2019.103539. Epub 2019 Nov 13. Comput Biol Med. 2020. PMID: 31765913 Review.
Cited by
- Integrative multi-omics analysis of chilling stress in pumpkin (Cucurbita moschata).
Li F, Liu B, Zhang H, Zhang J, Cai J, Cui J. Li F, et al. BMC Genomics. 2024 Nov 5;25(1):1042. doi: 10.1186/s12864-024-10939-2. BMC Genomics. 2024. PMID: 39501146 Free PMC article. - Olduvai domain expression downregulates mitochondrial pathways: implications for human brain evolution and neoteny.
Keeney JG, Astling D, Andries V, Vandepoele K, Anderson N, Davis JM, Lopert P, Vandenbussche J, Gevaert K, Staes A, Paukovich N, Vögeli B, Jones KL, van Roy F, Patel M, Sikela JM. Keeney JG, et al. bioRxiv [Preprint]. 2024 Oct 22:2024.10.21.619278. doi: 10.1101/2024.10.21.619278. bioRxiv. 2024. PMID: 39484454 Free PMC article. Preprint. - Decoding mutational hotspots in human disease through the gene modules governing thymic regulatory T cells.
Raposo AASF, Rosmaninho P, Silva SL, Paço S, Brazão ME, Godinho-Santos A, Tokunaga-Mizoro Y, Nunes-Cabaço H, Serra-Caetano A, Almeida ARM, Sousa AE. Raposo AASF, et al. Front Immunol. 2024 Oct 15;15:1458581. doi: 10.3389/fimmu.2024.1458581. eCollection 2024. Front Immunol. 2024. PMID: 39483472 Free PMC article. - How the extra X chromosome impairs the development of male fetal germ cells.
Lu Y, Qin M, He Q, Hua L, Qi X, Yang M, Guo Q, Liu X, Zhang Z, Xu F, Ding L, Wu Y, Zhang C, Zhai F, Liu Q, Li J, Yuan P, Shi X, Wang X, Zhao C, Lian Y, Li R, Wei Y, Yan L, Yuan P, Qiao J. Lu Y, et al. Nature. 2024 Oct 30. doi: 10.1038/s41586-024-08104-6. Online ahead of print. Nature. 2024. PMID: 39478217 - Inflammation impacts androgen receptor signaling in basal prostate stem cells through interleukin 1 receptor antagonist.
Cooper PO, Yang J, Wang HH, Broman MM, Jayasundara SM, Sahoo SS, Yan B, Awdalkreem GD, Cresswell GM, Wang L, Goossens E, Lanman NA, Doerge RW, Zheng F, Cheng L, Alqahtani S, Crist SA, Braun RE, Kazemian M, Jerde TJ, Ratliff TL. Cooper PO, et al. Commun Biol. 2024 Oct 25;7(1):1390. doi: 10.1038/s42003-024-07071-y. Commun Biol. 2024. PMID: 39455902 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources