High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli - PubMed (original) (raw)
High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli
Ekaterina Savitskaya et al. RNA Biol. 2013 May.
Abstract
In Escherichia coli, the acquisition of new CRISPR spacers is strongly stimulated by a priming interaction between a spacer in CRISPR RNA and a protospacer in foreign DNA. Priming also leads to a pronounced bias in DNA strand from which new spacers are selected. Here, ca. 200,000 spacers acquired during E. coli type I-E CRISPR/Cas-driven plasmid elimination were analyzed. Analysis of positions of plasmid protospacers from which newly acquired spacers have been derived is inconsistent with spacer acquisition machinery sliding along the target DNA as the primary mechanism responsible for strand bias during primed spacer acquisition. Most protospacers that served as donors of newly acquired spacers during primed spacer acquisition had an AAG protospacer adjacent motif, PAM. Yet, the introduction of multiple AAG sequences in the target DNA had no effect on the choice of protospacers used for adaptation, which again is inconsistent with the sliding mechanism. Despite a strong preference for an AAG PAM during CRISPR adaptation, the AAG (and CTT) triplets do not appear to be avoided in known E. coli phages. Likewise, PAM sequences are not avoided in Streptococcus thermophilus phages, indicating that CRISPR/Cas systems may not have been a strong factor in shaping host-virus interactions.
Keywords: CRISPR adaptation; CRISPR/Cas systems; Escherichia coli; bacteriophage; high-throughput sequencing.
Figures
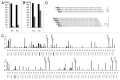
Figure 1. Experimental set-up to monitor CRISPR spacer acquisition. (A) At the top, the E. coli BL21 AI CRISPR 2.3 cassette is schematically presented, with repeats indicated as numbered gray rectangles. A leftward arrow indicates CRISPR promoter located in the leader sequence. The primers used to amplify the leader-proximal end of the cassette are shown (thick, not annealed parts of primers correspond to barcodes for high-throughput sequencing). Below, the structures of amplified DNA fragments expected in the absence (middle) or in the presence (bottom) of spacer acquisition are shown. (B) Results of PCR amplification using the primer set shown in (A) of DNA prepared from E. coli BL21 AI cultures transformed with plasmids pT7blue (lanes 1 and 3) or P1 and P2 plasmids (lanes 2 and 4) after an overnight growth at conditions of induction of plasmid-borne CRISPR/Cas components and in the absence of ampicillin needed to maintain pT7blue and its derivatives. The gray arrow indicates a PCR fragment arising from amplification of leader-proximal end of unexpanded CRSIRP 2.3 cassette; black arrow indicates a PCR fragment arising from amplification of CRISPR 2.3 cassette expanded by one spacer-repeat unit. Numbers at the left-hand side of the gel indicate the lengths (in bp) of DNA size markers. (C) Statistics of high-throughput sequencing of PCR amplification products extended by one spacer-repeat unit obtained with P1 and P2 samples.
Figure 2. Analysis of DNA strand and PAM preferences during primed and non-primed spacer acquisition. (A) Percentage of protospacers acquired from primed stand of the pT7blue vector in cells that lost the P1 and P2 plasmids are shown in black, percentage of protospacers from non-primed strand are shown in gray. (B) Percentage of AAG PAM sequences in protospacers acquired in cells that lost the P1 and P2 plasmids. Black bars show percentage of protospacers associated with AAG in the primed strand, gray-in the non-primed strand. (C) The distribution of non-AAG PAMs of primed strand (black bars) and non-primed strand (gray bars) protospacers in cells that lost P1 (“1”) and P2 (“2”) plasmids is presented. Non-AAG PAMs known to be functional in CRISPR interference are highlighted by bold typeface. The sum of percentage values for each plasmid/strand equals the total percentage of non-AAG PAMs in Figure 2B. (D) Two examples of imprecise spacer acquisition leading to appearance of non-AAG PAMs during primed acquisition. Spacer sequences are shown in regular typeface; numbers indicate their occurrences. The PAM sequences of corresponding protospacers are shown in bold typeface.
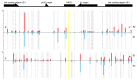
Figure 3. The distribution of donor protospacers in eliminated plasmids. The location of the ampicillin resistance bla gene, replication origins, and the multiple cloning site (MCS) of the pT7blue plasmid are schematically indicated at the top. Below, the two strands are shown separately as horizontal lines. The location of the priming protospacer is shown by yellow vertical line. The positions of AAG trinucleotides in each strand are shown by punctured vertical lines. For each strand, percentages of spacers corresponding to different protospacers are shown by red and blue vertical lines. Red lines correspond to primed and blue lines- to non-primed acquisition.
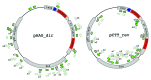
Figure 4. The distribution of donor protospacers acquired during primed spacer acquisition from plasmids containing multiple AAG sequence blocks. Two plasmids with opposing orientations of the priming protospacer (dark blue arrow) are schematically shown, with positions of poly AAG blocks highlighted in red. Protospacers corresponding to spacers acquired by cells that lost each plasmid are shown as green arrows. Protospacers on the outside originate from the coding strand of ampicillin-resistance gene bla. Protospacers on the inside originate from the opposite strand. Protospacers acquired from plasmids that lacked poly AAG tracks are shown in light green. Numbers indicate the number of times identical spacers have been observed.
Similar articles
- Avoidance of Trinucleotide Corresponding to Consensus Protospacer Adjacent Motif Controls the Efficiency of Prespacer Selection during Primed Adaptation.
Musharova O, Vyhovskyi D, Medvedeva S, Guzina J, Zhitnyuk Y, Djordjevic M, Severinov K, Savitskaya E. Musharova O, et al. mBio. 2018 Dec 4;9(6):e02169-18. doi: 10.1128/mBio.02169-18. mBio. 2018. PMID: 30514784 Free PMC article. - CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli.
Díez-Villaseñor C, Guzmán NM, Almendros C, García-Martínez J, Mojica FJ. Díez-Villaseñor C, et al. RNA Biol. 2013 May;10(5):792-802. doi: 10.4161/rna.24023. Epub 2013 Feb 27. RNA Biol. 2013. PMID: 23445770 Free PMC article. - CRISPR interference directs strand specific spacer acquisition.
Swarts DC, Mosterd C, van Passel MW, Brouns SJ. Swarts DC, et al. PLoS One. 2012;7(4):e35888. doi: 10.1371/journal.pone.0035888. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558257 Free PMC article. - CRISPR-Cas adaptation in Escherichia coli.
Mitić D, Bolt EL, Ivančić-Baće I. Mitić D, et al. Biosci Rep. 2023 Mar 31;43(3):BSR20221198. doi: 10.1042/BSR20221198. Biosci Rep. 2023. PMID: 36809461 Free PMC article. Review. - Detection of CRISPR adaptation.
Shiriaeva A, Fedorov I, Vyhovskyi D, Severinov K. Shiriaeva A, et al. Biochem Soc Trans. 2020 Feb 28;48(1):257-269. doi: 10.1042/BST20190662. Biochem Soc Trans. 2020. PMID: 32010936 Free PMC article. Review.
Cited by
- Type I-F CRISPR-Cas Distribution and Array Dynamics in Legionella pneumophila.
Deecker SR, Ensminger AW. Deecker SR, et al. G3 (Bethesda). 2020 Mar 5;10(3):1039-1050. doi: 10.1534/g3.119.400813. G3 (Bethesda). 2020. PMID: 31937548 Free PMC article. - Primed CRISPR adaptation in Escherichia coli cells does not depend on conformational changes in the Cascade effector complex detected in Vitro.
Krivoy A, Rutkauskas M, Kuznedelov K, Musharova O, Rouillon C, Severinov K, Seidel R. Krivoy A, et al. Nucleic Acids Res. 2018 May 4;46(8):4087-4098. doi: 10.1093/nar/gky219. Nucleic Acids Res. 2018. PMID: 29596641 Free PMC article. - Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer.
Richter C, Dy RL, McKenzie RE, Watson BN, Taylor C, Chang JT, McNeil MB, Staals RH, Fineran PC. Richter C, et al. Nucleic Acids Res. 2014 Jul;42(13):8516-26. doi: 10.1093/nar/gku527. Epub 2014 Jul 2. Nucleic Acids Res. 2014. PMID: 24990370 Free PMC article. - Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity.
Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Nuñez JK, et al. Nat Struct Mol Biol. 2014 Jun;21(6):528-34. doi: 10.1038/nsmb.2820. Epub 2014 May 4. Nat Struct Mol Biol. 2014. PMID: 24793649 Free PMC article. - CRISPR RNA binding and DNA target recognition by purified Cascade complexes from Escherichia coli.
Beloglazova N, Kuznedelov K, Flick R, Datsenko KA, Brown G, Popovic A, Lemak S, Semenova E, Severinov K, Yakunin AF. Beloglazova N, et al. Nucleic Acids Res. 2015 Jan;43(1):530-43. doi: 10.1093/nar/gku1285. Epub 2014 Dec 8. Nucleic Acids Res. 2015. PMID: 25488810 Free PMC article.
References
- Sharp PM. Molecular evolution of bacteriophages: evidence of selection against the recognition sites of host restriction enzymes. Mol Biol Evol. 1986;3:75–83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous