Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2 - PubMed (original) (raw)
. 2013 Jul;41(12):6347-59.
doi: 10.1093/nar/gkt315. Epub 2013 Apr 26.
Reinhild Wurm, Oleksandr Brener, Philipp Ellinger, Luitgard Nagel-Steger, Filipp Oesterhelt, Lutz Schmitt, Dieter Willbold, Rolf Wagner, Holger Gohlke, Sander H J Smits, Umit Pul
Affiliations
- PMID: 23625968
- PMCID: PMC3695520
- DOI: 10.1093/nar/gkt315
Double-strand DNA end-binding and sliding of the toroidal CRISPR-associated protein Csn2
Zihni Arslan et al. Nucleic Acids Res. 2013 Jul.
Abstract
The adaptive immunity of bacteria against foreign nucleic acids, mediated by CRISPR (clustered regularly interspaced short palindromic repeats), relies on the specific incorporation of short pieces of the invading foreign DNA into a special genomic locus, termed CRISPR array. The stored sequences (spacers) are subsequently used in the form of small RNAs (crRNAs) to interfere with the target nucleic acid. We explored the DNA-binding mechanism of the immunization protein Csn2 from the human pathogen Streptococcus agalactiae using different biochemical techniques, atomic force microscopic imaging and molecular dynamics simulations. The results demonstrate that the ring-shaped Csn2 tetramer binds DNA ends through its central hole and slides inward, likely by a screw motion along the helical path of the enclosed DNA. The presented data indicate an accessory function of Csn2 during integration of exogenous DNA by end-joining.
Figures
Figure 1.
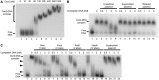
Electrophoretic mobility shift assays of a radiolabeled 155 bp DNA fragment with Csn2 either in the absence (A) or in the presence of competitor DNA (B and C) are presented. In each reaction 2 nM 32P-labeled DNA, 20 ng/µl heparin, and 10 mM CaCl2 were employed. (A) Titration of Csn2 in the range of 0 to 1 µM is shown. (B) Csn2 binding to the radiolabeled DNA fragment was competed with indicated amounts of 2915 bp unlabeled plasmid DNA either in _ScaI_-linearized (lanes 3–6), supercoiled (lanes 7–10) or in relaxed (lanes 11–14) form. The concentration of Csn2 was constant at 60 nM in lanes 2–14. Lanes 1 and 2 show the control reactions, performed either in the absence of Csn2 (lane 1) or in the absence of competitor DNA (lane 2). (C) The same competition experiment as in (B) but with _PvuII_-, _AvaII_-, or _HaeIII_-cleaved competitor plasmid. The numbers of cleavage sites of the different endonucleases are given in the brackets. The black arrowheads indicate intermediate Csn2-DNA complexes, resulting from decomposition of the fully occupied complexes.
Figure 2.
(A) Binding analyses of Csn2 in the presence and absence of EGTA and free DNA ends on 2% Tris-acetate agarose gel. In each lane 168 ng linear DNA and 7.2 mM CaCl2 were employed. The numbers above the lanes indicate the order of addition of streptavidin (2 µg), Csn2 (4.7 µg), or EGTA (14 mM) in a total volume of 14.4 µl. Lanes 2–5: Influence of EGTA on Csn2-DNA interaction is shown. Lanes 6–9: 168 ng of the end-biotinylated DNA fragment were incubated first with streptavidin to block the DNA ends. Lanes 10 and 11: Streptavidin was added after binding of Csn2. After separation of the complexes the agarose gel was stained with ethidium bromide. (B) Schematic presentation of the binding analysis, shown in (A).
Figure 3.
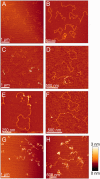
Representative AFM images of 5125 bp plasmid DNA in the absence or presence of Csn2 are shown. (A) and (B) show images of 1.3 nM relaxed and 1.3 nM linear plasmid DNA in the absence of Csn2; (C–F) images of equal amounts of relaxed and linear plasmid DNA (each 1.3 nM) incubated with 176 nM Csn2; (G) and (H) images of 2.6 nM linear plasmid DNA incubated with 800 nM Csn2. The relative color scale range is 0–3 nm in all images.
Figure 4.
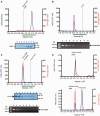
Analytical gel filtration analysis of Csn2 and Csn2-DNA complexes performed with a Superdex 200 PC 3.2/30 column is shown. Elution profiles of 20 µM Csn2 (A), 30 nM linearized pCR001 plasmid (B) and Csn2-DNA complexes (C) are shown. In (D) the elution profiles of 30 nM supercoiled pCR001 alone (upper part) or in the presence of 20 µM Csn2 (lower part) are shown. 100 µl fractions were collected starting at an elution volume of 0.8 ml. Aliquots of the fractions 1 to 8, indicated by the green lines below the elution profiles, were analyzed on 10% SDS gels (A, C) and on 1% agarose gels (B, C).
Figure 5.
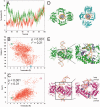
(A) Movement of Csn2 along the DNA determined from two independent MD simulations (red and green lines, respectively) over a time course of 70 ns. A translocation value of zero refers to Csn2 being centered on the DNA; translocation values >0 indicate a shift of Csn2 toward the termini of the DNA. (B) Coupled motions of translocation and rotation of Csn2 when moving along the DNA over the last 50 ns of the MD simulation. To determine the rotational motion, all conformations of the trajectories were aligned with respect to the phosphorous atoms of the DNA. (C) Coupling between DNA bending as determined by a kink angle and the translocation of Csn2 along the DNA over the last 50 ns of the MD simulation. (D) View along the DNA axis on Csn2-DNA conformations extracted from the MD trajectory at 50.3 ns (green) and 97.3 ns (cyan). The complexes were aligned with respect to the phosphorous atoms of the DNA and display a rotation of ∼75° of Csn2 during a translocation of ∼5.7 Å. In panel B, the respective data points are marked by circles. (E) Side view on Csn2-DNA complexes extracted from the MD trajectory at 50.3 ns (green) and 98.5 ns (magenta). The complexes display a kinking of the DNA by ∼40° during a translocation of Csn2 of ∼8.7 Å. In the close-up figures, helices H3 and loops β4-H2 are colored in blue, and Lys residues in these structural elements are depicted as sticks; unprimed labels mark helices and loops that belong to one dimer, primed labels mark objects that belong to the other dimer. In panel C, the respective data points are marked by circles.
Figure 6.
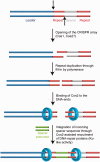
A model for the proposed function of Csn2 DNA end-binding in spacer integration is shown. The leader proximal repeat sequence serves as template for duplication of the repeat sequence (25,26,64), suggesting a cleavage at the leader-repeat and repeat-spacer borders (indicated by the dashed arrows). Complementary strand synthesis and binding of Csn2 to the DNA ends could assist in the integration of new spacer DNA fragments through holding the DNA ends together, while simultaneously recruiting DNA-repair proteins and protecting the double-stranded DNA breaks from exonucleolytic degradation.
Similar articles
- Crystal structure of clustered regularly interspaced short palindromic repeats (CRISPR)-associated Csn2 protein revealed Ca2+-dependent double-stranded DNA binding activity.
Nam KH, Kurinov I, Ke A. Nam KH, et al. J Biol Chem. 2011 Sep 2;286(35):30759-30768. doi: 10.1074/jbc.M111.256263. Epub 2011 Jun 21. J Biol Chem. 2011. PMID: 21697083 Free PMC article. - The crystal structure of the CRISPR-associated protein Csn2 from Streptococcus agalactiae.
Ellinger P, Arslan Z, Wurm R, Tschapek B, MacKenzie C, Pfeffer K, Panjikar S, Wagner R, Schmitt L, Gohlke H, Pul Ü, Smits SH. Ellinger P, et al. J Struct Biol. 2012 Jun;178(3):350-62. doi: 10.1016/j.jsb.2012.04.006. Epub 2012 Apr 17. J Struct Biol. 2012. PMID: 22531577 - Crystal structure of Streptococcus pyogenes Csn2 reveals calcium-dependent conformational changes in its tertiary and quaternary structure.
Koo Y, Jung DK, Bae E. Koo Y, et al. PLoS One. 2012;7(3):e33401. doi: 10.1371/journal.pone.0033401. Epub 2012 Mar 30. PLoS One. 2012. PMID: 22479393 Free PMC article. - Molecular Mechanisms of CRISPR-Cas Immunity in Bacteria.
Nussenzweig PM, Marraffini LA. Nussenzweig PM, et al. Annu Rev Genet. 2020 Nov 23;54:93-120. doi: 10.1146/annurev-genet-022120-112523. Epub 2020 Aug 28. Annu Rev Genet. 2020. PMID: 32857635 Review. - CRISPR-based adaptive and heritable immunity in prokaryotes.
van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. van der Oost J, et al. Trends Biochem Sci. 2009 Aug;34(8):401-7. doi: 10.1016/j.tibs.2009.05.002. Epub 2009 Jul 29. Trends Biochem Sci. 2009. PMID: 19646880 Review.
Cited by
- Global transcription of CRISPR loci in the human oral cavity.
Lum AG, Ly M, Santiago-Rodriguez TM, Naidu M, Boehm TK, Pride DT. Lum AG, et al. BMC Genomics. 2015 May 21;16(1):401. doi: 10.1186/s12864-015-1615-0. BMC Genomics. 2015. PMID: 25994215 Free PMC article. - Molecular organization of the type II-A CRISPR adaptation module and its interaction with Cas9 via Csn2.
Ka D, Jang DM, Han BW, Bae E. Ka D, et al. Nucleic Acids Res. 2018 Oct 12;46(18):9805-9815. doi: 10.1093/nar/gky702. Nucleic Acids Res. 2018. PMID: 30102386 Free PMC article. - Digging into the lesser-known aspects of CRISPR biology.
Guzmán NM, Esquerra-Ruvira B, Mojica FJM. Guzmán NM, et al. Int Microbiol. 2021 Nov;24(4):473-498. doi: 10.1007/s10123-021-00208-7. Epub 2021 Sep 6. Int Microbiol. 2021. PMID: 34487299 Free PMC article. Review. - CRISPR DNA elements controlling site-specific spacer integration and proper repeat length by a Type II CRISPR-Cas system.
Kim JG, Garrett S, Wei Y, Graveley BR, Terns MP. Kim JG, et al. Nucleic Acids Res. 2019 Sep 19;47(16):8632-8648. doi: 10.1093/nar/gkz677. Nucleic Acids Res. 2019. PMID: 31392984 Free PMC article. - Quantifying length-dependent DNA end-binding by nucleoproteins.
Vo T, Albrecht AV, Wilson WD, Poon GMK. Vo T, et al. Biophys Chem. 2019 Aug;251:106177. doi: 10.1016/j.bpc.2019.106177. Epub 2019 Apr 30. Biophys Chem. 2019. PMID: 31102748 Free PMC article.
References
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45:273–297. - PubMed
- Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol. Chem. 2011;392:277–289. - PubMed
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. - PubMed
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources