phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data - PubMed (original) (raw)
phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data
Paul J McMurdie et al. PLoS One. 2013.
Abstract
Background: the analysis of microbial communities through dna sequencing brings many challenges: the integration of different types of data with methods from ecology, genetics, phylogenetics, multivariate statistics, visualization and testing. With the increased breadth of experimental designs now being pursued, project-specific statistical analyses are often needed, and these analyses are often difficult (or impossible) for peer researchers to independently reproduce. The vast majority of the requisite tools for performing these analyses reproducibly are already implemented in R and its extensions (packages), but with limited support for high throughput microbiome census data.
Results: Here we describe a software project, phyloseq, dedicated to the object-oriented representation and analysis of microbiome census data in R. It supports importing data from a variety of common formats, as well as many analysis techniques. These include calibration, filtering, subsetting, agglomeration, multi-table comparisons, diversity analysis, parallelized Fast UniFrac, ordination methods, and production of publication-quality graphics; all in a manner that is easy to document, share, and modify. We show how to apply functions from other R packages to phyloseq-represented data, illustrating the availability of a large number of open source analysis techniques. We discuss the use of phyloseq with tools for reproducible research, a practice common in other fields but still rare in the analysis of highly parallel microbiome census data. We have made available all of the materials necessary to completely reproduce the analysis and figures included in this article, an example of best practices for reproducible research.
Conclusions: The phyloseq project for R is a new open-source software package, freely available on the web from both GitHub and Bioconductor.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
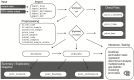
Figure 1. Example of a phylogenetic sequencing workflow.
A diagram of an experimental and analysis workflow for amplicon or shotgun phylogenetic sequencing. The intended role for phyloseq is indicated.
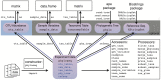
Figure 2. Analysis workflow using phyloseq.
The workflow starts with the results of OTU clustering and independently-measured sample data (Input, top left), and ends at various analytic procedures available in R for inference and validation. In between are key functions for preprocessing and graphics. Rounded rectangles and diamond shapes represent functions and data objects, respectively, further described in Figure 3.
Figure 3. The “phyloseq” class.
The phyloseq class is an experiment-level data storage class defined by the phyloseq package for representing phylogenetic sequencing data. Most functions in the phyloseq package expect an instance of this class as their primary argument. See the phyloseq manual for a complete list of functions.
Figure 4. Graphic functions of the phyloseq package.
The phyloseq class is an experiment-level data storage class defined by the phyloseq package for representing phylogenetic sequencing data. Most functions in the phyloseq package expect an instance of this class as their primary argument. See the phyloseq manual The Global Patterns and Enterotypes datasets are included with the phyloseq package. The Global Patterns data was preprocessed such that each sample was transformed to the same total read depth, and OTUs were trimmed that were not observed at least 3 times in 20% of samples or had a coefficient of variation ≤ 3.0 across all samples. For the plot_tree and plot_bar subplots, only the Bacteroidetes phylum is shown. Each subplot title indicates the plot function that produced it. Complete details for reproducing this figure are provided in File S2. All of these functions return a ggplot object that can be further customized/modified by tools in the ggplot2 package . See additional descriptions of each function in the body text, and at the phyloseq homepage .
Figure 5. plot_ordination display methods included in phyloseq.
Each panel uses a “Bacteroidetes-only” subset of the preprocessed “Global Patterns” dataset that was also used in Figure 4. The coordinates are derived from an unconstrained correspondence analysis . Different panels illustrate different displays of the ordination results using the type argument to the plot_ordination function. (Top Left) Example of a samples-only display, with the “SampleType” mapped to the color aesthetic, and a filled-polygon layer to emphasize plot regions where sample types co-occur. (Top Left Insert) A “scree” plot of the eigenvalues associated with each axis, which indicates the proportion of total variability represented in each axis. (Top Right) Biplot representation in which samples and OTUs ordination results are overlaid. Clumps of OTUs appear to co-occur with different sample types, and some correlation with taxonomic phylum is also evident. (Middle) An OTUs-only plot that has been faceted (separated into panels) by class, with a two-dimensional density estimate overlain in blue. This view shows clearly a lack of association between the Sphingobacteria and Flavobacteria classes with fecal samples, which appear to be enriched in a subset of the Bacteroidia (relative to other OTUs in this Bacteroidetes-only dataset). Meanwhile, subsets of Bacteroidia appear to be enriched within multiple sample types. (Bottom) The “split” type for this graphic, in which both samples-only and OTUs-only plots are created, and shown side-by-side with one legend and shared vertical axis. Both the “biplot” and “split” options allow dual projections of both OTU- and sample-space.
Similar articles
- phylogeo: an R package for geographic analysis and visualization of microbiome data.
Charlop-Powers Z, Brady SF. Charlop-Powers Z, et al. Bioinformatics. 2015 Sep 1;31(17):2909-11. doi: 10.1093/bioinformatics/btv269. Epub 2015 Apr 25. Bioinformatics. 2015. PMID: 25913208 Free PMC article. - Shiny-phyloseq: Web application for interactive microbiome analysis with provenance tracking.
McMurdie PJ, Holmes S. McMurdie PJ, et al. Bioinformatics. 2015 Jan 15;31(2):282-3. doi: 10.1093/bioinformatics/btu616. Epub 2014 Sep 26. Bioinformatics. 2015. PMID: 25262154 Free PMC article. - The best practice for microbiome analysis using R.
Wen T, Niu G, Chen T, Shen Q, Yuan J, Liu YX. Wen T, et al. Protein Cell. 2023 Oct 25;14(10):713-725. doi: 10.1093/procel/pwad024. Protein Cell. 2023. PMID: 37128855 Free PMC article. - A review of software for analyzing molecular sequences.
Nilakanta H, Drews KL, Firrell S, Foulkes MA, Jablonski KA. Nilakanta H, et al. BMC Res Notes. 2014 Nov 24;7:830. doi: 10.1186/1756-0500-7-830. BMC Res Notes. 2014. PMID: 25421430 Free PMC article. Review. - Practical considerations for sampling and data analysis in contemporary metagenomics-based environmental studies.
Staley C, Sadowsky MJ. Staley C, et al. J Microbiol Methods. 2018 Nov;154:14-18. doi: 10.1016/j.mimet.2018.09.020. Epub 2018 Oct 1. J Microbiol Methods. 2018. PMID: 30287354 Review.
Cited by
- Taxonomic and metabolic characterisation of biofilms colonising Roman stuccoes at Baia's thermal baths and restoration strategies.
De Luca D, Piredda R, Scamardella S, Martelli Castaldi M, Troisi J, Lombardi M, De Castro O, Cennamo P. De Luca D, et al. Sci Rep. 2024 Nov 1;14(1):26290. doi: 10.1038/s41598-024-76637-x. Sci Rep. 2024. PMID: 39487240 Free PMC article. - A cross-systems primer for synthetic microbial communities.
Mehlferber EC, Arnault G, Joshi B, Partida-Martinez LP, Patras KA, Simonin M, Koskella B. Mehlferber EC, et al. Nat Microbiol. 2024 Nov;9(11):2765-2773. doi: 10.1038/s41564-024-01827-2. Epub 2024 Oct 30. Nat Microbiol. 2024. PMID: 39478083 Free PMC article. Review. - Microbial community dynamics in blood, faeces and oral secretions of neotropical bats in Casanare, Colombia.
Luna N, Páez-Triana L, Ramírez AL, Muñoz M, Goméz M, Medina JE, Urbano P, Barragán K, Ariza C, Martínez D, Hernández C, Patiño LH, Ramirez JD. Luna N, et al. Sci Rep. 2024 Oct 28;14(1):25808. doi: 10.1038/s41598-024-77090-6. Sci Rep. 2024. PMID: 39468253 Free PMC article. - Deep Soil Layers of Drought-Exposed Forests Harbor Poorly Known Bacterial and Fungal Communities.
Frey B, Walthert L, Perez-Mon C, Stierli B, Köchli R, Dharmarajah A, Brunner I. Frey B, et al. Front Microbiol. 2021 May 7;12:674160. doi: 10.3389/fmicb.2021.674160. eCollection 2021. Front Microbiol. 2021. PMID: 34025630 Free PMC article. - The intestinal microbiota and metabolites in patients with anorexia nervosa.
Prochazkova P, Roubalova R, Dvorak J, Kreisinger J, Hill M, Tlaskalova-Hogenova H, Tomasova P, Pelantova H, Cermakova M, Kuzma M, Bulant J, Bilej M, Smitka K, Lambertova A, Holanova P, Papezova H. Prochazkova P, et al. Gut Microbes. 2021 Jan-Dec;13(1):1-25. doi: 10.1080/19490976.2021.1902771. Gut Microbes. 2021. PMID: 33779487 Free PMC article.
References
- Metzker ML (2010) Sequencing technologies - the next generation. Nature Reviews Genetics 11: 31–46. - PubMed
- Pace NR (1997) A molecular view of microbial diversity and the biosphere. Science 276: 734–740. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources