One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering - PubMed (original) (raw)
One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering
Haoyi Wang et al. Cell. 2013.
Abstract
Mice carrying mutations in multiple genes are traditionally generated by sequential recombination in embryonic stem cells and/or time-consuming intercrossing of mice with a single mutation. The CRISPR/Cas system has been adapted as an efficient gene-targeting technology with the potential for multiplexed genome editing. We demonstrate that CRISPR/Cas-mediated gene editing allows the simultaneous disruption of five genes (Tet1, 2, 3, Sry, Uty--8 alleles) in mouse embryonic stem (ES) cells with high efficiency. Coinjection of Cas9 mRNA and single-guide RNAs (sgRNAs) targeting Tet1 and Tet2 into zygotes generated mice with biallelic mutations in both genes with an efficiency of 80%. Finally, we show that coinjection of Cas9 mRNA/sgRNAs with mutant oligos generated precise point mutations simultaneously in two target genes. Thus, the CRISPR/Cas system allows the one-step generation of animals carrying mutations in multiple genes, an approach that will greatly accelerate the in vivo study of functionally redundant genes and of epistatic gene interactions.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
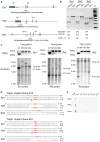
Figure 1. Multiplexed Gene Targeting in mouse ES cells
(A) Schematic of the Cas9/sgRNA-targeting sites in Tet1, 2, and 3. The sgRNA-targeting sequence is underlined, and the protospacer-adjacent motif (PAM) sequence is labeled in green. The restriction sites at the target regions are bold and capitalized. Restriction enzymes used for RFLP and Southern blot analysis are shown, and the Southern blot probes are shown as orange boxes. (B) Surveyor assay for Cas9-mediated cleavage at Tet1, 2, and 3 loci in ES cells. (C) Genotyping of triple-targeted ES cells, clones 51, 52, and 53 are shown. Upper: RFLP analysis. Tet1 PCR products were digested with SacI, Tet2 PCR products were digested with EcoRV, and Tet3 PCR products were digested with XhoI. Lower: Southern blot analysis. For the Tet1 locus, SacI digested genomic DNA was hybridized with a 5′ probe. Expected fragment size: WT = 5.8 kb, TM (targeted mutation) = 6.4 kb. For the Tet2 locus, SacI, and EcoRV double-digested genomic DNA was hybridized with a 3′ probe. Expected fragment size: WT = 4.3 kb, TM = 5.6 kb. For the Tet3 locus, BamHI and XhoI double-digested genomic DNA was hybridized with a 5′ probe. Expected fragment size: WT = 3.2 kb, TM = 8.1 kb. (D) The sequence of six mutant alleles in triple-targeted ES cell clone 14 and 41. PAM sequence is labeled in red. (E) Analysis of 5hmC levels in DNA isolated from triple-targeted ES cell clones by dot blot assay using anti-5hmC antibody. A previously characterized DKO clone derived using traditional method is used as a control. See also Figure S1.
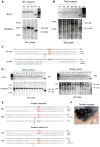
Figure 2. Single- and Double-Gene Targeting In Vivo by Injection into Fertilized Eggs
(A) Genotyping of Tet1 single-targeted mice. (B) Upper: genotyping of Tet2 single-targeted mice. RFLP analysis; lower: Southern blot analysis. (C) The sequence of both alleles of targeted gene in Tet1 biallelic mutant mouse 2 and Tet2 biallelic mutant mouse 4. (D) Genotyping of Tet1/Tet2 double-mutant mice. Analysis of mice 1 to 12 is shown. Upper: RFLP analysis; lower: southern blot analysis. The Tet1 locus is displayed on the left and the Tet2 locus on the right. (E) The sequence of four mutant alleles from double-mutant mouse 9 and 10. PAM sequences are labeled in red. (F) Three-week-old double-mutant mice. All RFLP and Southern digestions and probes are the same as those used in Figure 1. See also Figures S2 and S3.
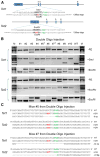
Figure 3. Multiplexed HDR-Mediated Genome Editing In Vivo
(A) Schematic of the oligo-targeting sites at Tet1 and Tet2 loci. The sgRNA-targeting sequence is underlined, and the PAM sequence is labeled in green. Oligo targeting each gene is shown under the target site, with 2 bp changes labeled in red. Restriction enzyme sites used for RFLP analysis are bold and capitalized. (B) RFLP analysis of double oligo injection mice with HDR-mediated targeting at the Tet1 and Tet2 loci. (C) The sequences of both alleles of Tet1 and Tet2 in mouse 5 and 7 show simultaneously HDR-mediated targeting at one allele or two alleles of each gene, and NHEJ-mediated disruption at the other alleles. See also Figure S4.
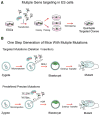
Figure 4. Mutiplexed Genome Editing in ES Cells and Mouse
(A) Multiple gene targeting in ES cells. (B) One-step generation of mice with multiple mutations. Upper: multiple targeted mutations with random indels introduced through NHEJ. Lower: multiple predefined mutations introduced through HDR-mediated repair.
Similar articles
- One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering.
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. Yang H, et al. Cell. 2013 Sep 12;154(6):1370-9. doi: 10.1016/j.cell.2013.08.022. Epub 2013 Aug 29. Cell. 2013. PMID: 23992847 Free PMC article. - Generating genetically modified mice using CRISPR/Cas-mediated genome engineering.
Yang H, Wang H, Jaenisch R. Yang H, et al. Nat Protoc. 2014 Aug;9(8):1956-68. doi: 10.1038/nprot.2014.134. Epub 2014 Jul 24. Nat Protoc. 2014. PMID: 25058643 - CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice.
Oji A, Noda T, Fujihara Y, Miyata H, Kim YJ, Muto M, Nozawa K, Matsumura T, Isotani A, Ikawa M. Oji A, et al. Sci Rep. 2016 Aug 17;6:31666. doi: 10.1038/srep31666. Sci Rep. 2016. PMID: 27530713 Free PMC article. - The new CRISPR-Cas system: RNA-guided genome engineering to efficiently produce any desired genetic alteration in animals.
Seruggia D, Montoliu L. Seruggia D, et al. Transgenic Res. 2014 Oct;23(5):707-16. doi: 10.1007/s11248-014-9823-y. Epub 2014 Aug 6. Transgenic Res. 2014. PMID: 25092533 Review. - A mouse geneticist's practical guide to CRISPR applications.
Singh P, Schimenti JC, Bolcun-Filas E. Singh P, et al. Genetics. 2015 Jan;199(1):1-15. doi: 10.1534/genetics.114.169771. Epub 2014 Sep 29. Genetics. 2015. PMID: 25271304 Free PMC article. Review.
Cited by
- Opposing roles of E3 ligases TRIM23 and TRIM21 in regulation of ion channel ANO1 protein levels.
Cao X, Zhou Z, Tian Y, Liu Z, Cheng KO, Chen X, Hu W, Wong YM, Li X, Zhang H, Hu R, Huang P. Cao X, et al. J Biol Chem. 2021 Jan-Jun;296:100738. doi: 10.1016/j.jbc.2021.100738. Epub 2021 May 3. J Biol Chem. 2021. PMID: 33957127 Free PMC article. - Histone methyltransferase Ash1L mediates activity-dependent repression of neurexin-1α.
Zhu Τ, Liang C, Li D, Tian M, Liu S, Gao G, Guan JS. Zhu Τ, et al. Sci Rep. 2016 May 27;6:26597. doi: 10.1038/srep26597. Sci Rep. 2016. PMID: 27229316 Free PMC article. - Dephosphorylated parafibromin is a transcriptional coactivator of the Wnt/Hedgehog/Notch pathways.
Kikuchi I, Takahashi-Kanemitsu A, Sakiyama N, Tang C, Tang PJ, Noda S, Nakao K, Kassai H, Sato T, Aiba A, Hatakeyama M. Kikuchi I, et al. Nat Commun. 2016 Sep 21;7:12887. doi: 10.1038/ncomms12887. Nat Commun. 2016. PMID: 27650679 Free PMC article. - Continuous live imaging reveals a subtle pathological alteration with cell behaviors in congenital heart malformation.
Li X, Yue Y, Zhang Y, Liao Y, Wang Q, Bian Y, Na J, He A. Li X, et al. Fundam Res. 2021 Dec 3;2(1):14-22. doi: 10.1016/j.fmre.2021.11.025. eCollection 2022 Jan. Fundam Res. 2021. PMID: 38933910 Free PMC article. - Prefrontal cortex and the dysconnectivity hypothesis of schizophrenia.
Zhou Y, Fan L, Qiu C, Jiang T. Zhou Y, et al. Neurosci Bull. 2015 Apr;31(2):207-19. doi: 10.1007/s12264-014-1502-8. Epub 2015 Mar 11. Neurosci Bull. 2015. PMID: 25761914 Free PMC article. Review.
References
- Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1846. - PubMed
- Capecchi MR. Gene targeting in mice: functional analysis of the mammalian genome for the twenty-first century. Nat Rev Genet. 2005;6:507–512. - PubMed
- Carroll D, Beumer KJ, Morton JJ, Bozas A, Trautman JK. Gene targeting in Drosophila and Caenorhabditis elegans with zinc-finger nucleases. Methods Mol Biol. 2008;435:63–77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01-CA084198/CA/NCI NIH HHS/United States
- R37 HD045022/HD/NICHD NIH HHS/United States
- DP1 MH100706/MH/NIMH NIH HHS/United States
- R01 CA084198/CA/NCI NIH HHS/United States
- R37-HD045022/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases