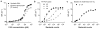A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria - PubMed (original) (raw)
A magneto-DNA nanoparticle system for rapid detection and phenotyping of bacteria
Hyun Jung Chung et al. Nat Nanotechnol. 2013 May.
Abstract
So far, although various diagnostic approaches for pathogen detection have been proposed, most are too expensive, lengthy or limited in specificity for clinical use. Nanoparticle systems with unique material properties, however, circumvent these problems and offer improved accuracy over current methods. Here, we present novel magneto-DNA probes capable of rapid and specific profiling of pathogens directly in clinical samples. A nanoparticle hybridization assay, involving ubiquitous and specific probes that target bacterial 16S rRNAs, was designed to detect amplified target DNAs using a miniaturized NMR device. Ultimately, the magneto-DNA platform will allow both universal and specific detection of various clinically relevant bacterial species, with sensitivity down to single bacteria. Furthermore, the assay is robust and rapid, simultaneously diagnosing a panel of 13 bacterial species in clinical specimens within 2 h. The generic platform described could be used to rapidly identify and phenotype pathogens for a variety of applications.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
Figure 1. Magneto-DNA assay for the detection of bacterial 16S rRNA
a, Schematic of the assay procedure. Total RNA is extracted from the specimen, and the 16S rRNA is amplified by asymmetric real time-PCR. Single-strand DNA of the amplified product is then captured by beads conjugated to capture probes, before hybridising with MNPs to form a magnetic sandwich complex. Samples are subsequently analyzed using a _μ_NMR system. b, Hybridised probe complexes, as observed by transmission electron microscopy (left, size bar 100 nm), scanning electron microscopy (center, size bar 300 nm), and atomic force microscopy (right, size bar 100 nm).
Figure 2. Detection sensitivity of the magneto-DNA system
a, Serial dilutions of synthetic DNA or bacteria-derived DNA were used as detection targets. Bacteria-derived DNA molecules were obtained via asymmetric RT-PCR of S. aureus 16S rRNA (35 cycles); synthetic DNA had the same sequence as bacteria-derived DNA. The detection limit was ~ 0.5 pM. b, Bacterial detection by the magneto-DNA assay. Samples with varying numbers of S. aureus were used. Total RNA was extracted and target sequences were amplified by 30, 35, and 40 cycles of RT-PCR. Amplified target DNA were detected using the probe set in a. The observed detection limit was a single bacterium, and the dynamic range of detection could be controlled by changing the PCR cycle number. c, Bacterial detection in blood. Serial dilutions of E. coli were spiked into human blood and processed by first lysing the red blood cells and then extracting the RNA using the same procedure as described above. All experiments were done in triplicate. All Δ_R_2 values were obtained by subtracting the relaxation rate values of the hybridised probe complexes in the presence of target DNA (_R_2,target), with the relaxation rate values of the beads alone (_R_2,control). Data are expressed as mean ± SD.
Figure 3. Universal detection of bacteria using the magneto-DNA system
a, Sequences of universal probes targeting a conserved region of bacterial 16S rRNA. Two probe sequences with a single-base difference were blended and used for capture. (R, Y) = (A, T) for Staphylococcus, Escherichia, Pseudomonas, Klebsiella, Enterobacter, Haemophilus, Stenotrophomonas; (R, Y) = (G, C) for Streptococcus, Enterococcus, Acinetobacter, Proteus, Lactobacillus. b, Thirteen different bacterial species could each be detected using the universal probes. c, Mixtures containing different bacterial types were detected by the universal probes. The observed Δ_R_2 values were consistent with average Δ_R_2 value (dotted line) from single species. Data are expressed as mean ± SD. All samples for the assay were prepared in triplicate.
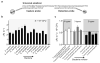
Figure 4. Differential detection using the magneto-DNA system
Probes targeting hypervariable regions of bacterial 16S rRNA sequences were used to specifically detect various bacterial types. RNA was extracted from bacterial cultures, amplified by asymmetric RT-PCR (35 cycles) using specific primers for each species, and detected using the corresponding probe conjugates. a, Probes specific for Staphylococcus were used for detecting S. aureus (DNA amount equivalent to 50,000 CFU). Target DNA from other bacterial species were added as controls to test off-target binding of the probes. b, Relaxation rates for differential detection of various bacterial types. Note the high specific signals and low background noise against other bacteria. Data are expressed as mean ± SD. All samples for the assay were prepared in triplicate. c,d, Heat maps comparing the specificity of the magneto-DNA assay with that of qPCR. Specificities in c were based on Δ_R_2 values from the magneto-DNA assay shown in b. Specificities in d are relative target amounts obtained from qPCR in Figure S5. Significant signals were marked as positive: positive signals for specific target bacteria were regarded as ‘true-positive’, while positive signals from non-targeted samples were classed as ‘false-positive’.
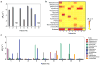
Figure 5. Diagnosis of clinical samples
Detection of pathogens by the magneto-DNA nanoparticle system using universal (a), and specific (c) probes for each bacteria type. b, Heatmap of obtained Δ_R_2 values for universal and specific detection. Clinical specimens (300 _μ_l for each sample) were processed to extract total RNA. This was followed by asymmetric RT-PCR (35 cycles) with universal and specific primers for all bacterial types. The PCR products (equivalent to 0.3 _μ_l volume of sample) were magnetically labeled and detected by _μ_NMR. a, Two of the 9 clinical samples tested negative, which correlated well with standard culture results. The other 7 samples were positive. ND represents samples with no pathogens detected. c, Identification of pathogen types within each sample. Note that some samples were infected with more than one pathogen, and the identified bacterial types correlated well with standard culture (see also Supplementary Table S3). Data are expressed as mean ± SD. All samples for the assay were prepared in triplicate.
Comment in
- Magnetic sensors: nanoparticles detect infection.
McNally A. McNally A. Nat Nanotechnol. 2013 May;8(5):315-6. doi: 10.1038/nnano.2013.76. Nat Nanotechnol. 2013. PMID: 23648737 No abstract available.
Similar articles
- Electrical Signal Reporter, Pore-Forming Protein, for Rapid, Miniaturized, and Universal Identification of Microorganisms.
Wan Y, Song F, Wang G, Liu H, An M, Wang A, Wu X, Ma C, Wang N. Wan Y, et al. Anal Chem. 2018 Aug 21;90(16):9853-9858. doi: 10.1021/acs.analchem.8b01933. Epub 2018 Aug 1. Anal Chem. 2018. PMID: 30024735 - Magnetic sensors: nanoparticles detect infection.
McNally A. McNally A. Nat Nanotechnol. 2013 May;8(5):315-6. doi: 10.1038/nnano.2013.76. Nat Nanotechnol. 2013. PMID: 23648737 No abstract available. - PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid.
Greisen K, Loeffelholz M, Purohit A, Leong D. Greisen K, et al. J Clin Microbiol. 1994 Feb;32(2):335-51. doi: 10.1128/jcm.32.2.335-351.1994. J Clin Microbiol. 1994. PMID: 7512093 Free PMC article. - Assessment of the microbiota of a mixed infection of the tongue using phenotypic and genotypic methods simultaneously and a review of the literature.
Veloo AC, Schepers RH, Welling GW, Degener JE. Veloo AC, et al. Anaerobe. 2011 Apr;17(2):47-51. doi: 10.1016/j.anaerobe.2011.03.005. Epub 2011 Mar 31. Anaerobe. 2011. PMID: 21458578 Review. - Characterization of microbial pathogens by DNA microarrays.
Huyghe A, Francois P, Schrenzel J. Huyghe A, et al. Infect Genet Evol. 2009 Sep;9(5):987-95. doi: 10.1016/j.meegid.2008.10.016. Epub 2008 Nov 17. Infect Genet Evol. 2009. PMID: 19061975 Free PMC article. Review. No abstract available.
Cited by
- Frequency Mixing Magnetic Detection Setup Employing Permanent Ring Magnets as a Static Offset Field Source.
Pourshahidi AM, Achtsnicht S, Offenhäusser A, Krause HJ. Pourshahidi AM, et al. Sensors (Basel). 2022 Nov 14;22(22):8776. doi: 10.3390/s22228776. Sensors (Basel). 2022. PMID: 36433383 Free PMC article. - Homogeneous Biosensing Based on Magnetic Particle Labels.
Schrittwieser S, Pelaz B, Parak WJ, Lentijo-Mozo S, Soulantica K, Dieckhoff J, Ludwig F, Guenther A, Tschöpe A, Schotter J. Schrittwieser S, et al. Sensors (Basel). 2016 Jun 6;16(6):828. doi: 10.3390/s16060828. Sensors (Basel). 2016. PMID: 27275824 Free PMC article. Review. - Click Chemistry-Mediated Nanosensors for Biochemical Assays.
Chen Y, Xianyu Y, Wu J, Yin B, Jiang X. Chen Y, et al. Theranostics. 2016 Apr 28;6(7):969-85. doi: 10.7150/thno.14856. eCollection 2016. Theranostics. 2016. PMID: 27217831 Free PMC article. Review. - Multiplexed microRNA Expression Profiling by Combined Asymmetric PCR and Label-Free Detection using Silicon Photonic Sensor Arrays.
Graybill RM, Cardenosa-Rubio MC, Yang H, Johnson MD, Bailey RC. Graybill RM, et al. Anal Methods. 2018 Apr 14;10(14):1618-1623. doi: 10.1039/C8AY00190A. Epub 2018 Mar 23. Anal Methods. 2018. PMID: 30275912 Free PMC article. - Increased sensitivity of enterotoxigenic Escherichia coli detection in stool samples using oligonucleotide immobilized-magnetic nanoparticles.
Jangpatarapongsa K, Saimuang K, Polpanich D, Thiramanas R, Techakasikornpanich M, Yudech P, Paripurana V, Leepiyasakulchai C, Tangboriboonrat P. Jangpatarapongsa K, et al. Biotechnol Rep (Amst). 2021 Sep 24;32:e00677. doi: 10.1016/j.btre.2021.e00677. eCollection 2021 Dec. Biotechnol Rep (Amst). 2021. PMID: 34631437 Free PMC article.
References
- Allegranzi B, et al. Burden of endemic health-care-associated infection in developing countries: systematic review and meta-analysis. The Lancet. 2011;377:228–241. - PubMed
- Polin RA, et al. Epidemiology and Diagnosis of Health Care–Associated Infections in the NICU. Pediatrics. 2012;129:e1104–e1109. - PubMed
- Klompas M, Yokoe DS, Weinstein RA. Automated surveillance of health care–associated infections. Clin Infect Dis. 2009;48:1268–1275. - PubMed
- Loman NJ, et al. Performance comparison of benchtop high-throughput sequencing platforms. Nat Biotechnol. 2012;30:434–439. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01HL113156/HL/NHLBI NIH HHS/United States
- R01EB010011/EB/NIBIB NIH HHS/United States
- HHSN268201000044C/HL/NHLBI NIH HHS/United States
- 268201000044C/PHS HHS/United States
- R01EB004626/EB/NIBIB NIH HHS/United States
- R01 EB010011/EB/NIBIB NIH HHS/United States
- R01 EB004626/EB/NIBIB NIH HHS/United States
- R01 HL113156/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources