The comprehensive antibiotic resistance database - PubMed (original) (raw)
. 2013 Jul;57(7):3348-57.
doi: 10.1128/AAC.00419-13. Epub 2013 May 6.
Nicholas Waglechner, Fazmin Nizam, Austin Yan, Marisa A Azad, Alison J Baylay, Kirandeep Bhullar, Marc J Canova, Gianfranco De Pascale, Linda Ejim, Lindsay Kalan, Andrew M King, Kalinka Koteva, Mariya Morar, Michael R Mulvey, Jonathan S O'Brien, Andrew C Pawlowski, Laura J V Piddock, Peter Spanogiannopoulos, Arlene D Sutherland, Irene Tang, Patricia L Taylor, Maulik Thaker, Wenliang Wang, Marie Yan, Tennison Yu, Gerard D Wright
Affiliations
- PMID: 23650175
- PMCID: PMC3697360
- DOI: 10.1128/AAC.00419-13
The comprehensive antibiotic resistance database
Andrew G McArthur et al. Antimicrob Agents Chemother. 2013 Jul.
Abstract
The field of antibiotic drug discovery and the monitoring of new antibiotic resistance elements have yet to fully exploit the power of the genome revolution. Despite the fact that the first genomes sequenced of free living organisms were those of bacteria, there have been few specialized bioinformatic tools developed to mine the growing amount of genomic data associated with pathogens. In particular, there are few tools to study the genetics and genomics of antibiotic resistance and how it impacts bacterial populations, ecology, and the clinic. We have initiated development of such tools in the form of the Comprehensive Antibiotic Research Database (CARD; http://arpcard.mcmaster.ca). The CARD integrates disparate molecular and sequence data, provides a unique organizing principle in the form of the Antibiotic Resistance Ontology (ARO), and can quickly identify putative antibiotic resistance genes in new unannotated genome sequences. This unique platform provides an informatic tool that bridges antibiotic resistance concerns in health care, agriculture, and the environment.
Figures
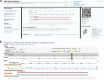
Fig 1
Presentation of the Staphylococcus aureus blaZ ß-lactamase gene in the CARD. (A) The gene's Web page in the CARD, providing annotation, accession, source information, ontological classification (SO, ARO, GO), and associated molecular features (mRNA, polypeptide, coding sequence [CDS]). (B) Dynamic browsing and analysis of the blaZ gene using the Web-based genome visualization and analysis tool GBrowse.
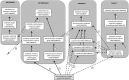
Fig 2
Classification of aminocoumarin-resistant gyrase B in the Antibiotic Resistance Ontology (ARO), illustrating the use of ontological relationships to describe knowledge about the gene (see Table 3). The is_a relationships are depicted by solid arrows labeled with “i” and generally denote classification hierarchies within the major branches of the ARO (mechanism, determinant, antibiotic, target), while dashed arrows labeled with “p” reflect part_of relationships between genes and mechanisms. Dashed arrows labeled with “d” depict derived_from relationships between antibiotic-sensitive precursors and antibiotic-resistant forms of the gene, while those labeled with “t” reflect targeted_by relationships between antibiotic-sensitive forms and antibiotic molecules. Dashed arrows labeled with “r” depict confers_resistance relationships between antibiotic resistance genes and antibiotic molecules. Asterisks denote a derived_from relationship between antibiotic-resistant and -sensitive DNA topoisomerase subunits.
Fig 3
Organization of aminoglycoside antibiotics (blue), their target (green), and aminoglycoside resistance genes (red) in the CARD's Antibiotic Resistance Ontology, illustrating the diversity of genes providing resistance to single or multiple aminoglycosides. Nodes represent ontology terms, while edges represent relationships between ontology terms.
Fig 4
An ontology term Web page for tetracycline resistance gene tetX (ARO:3000205) in the CARD, providing descriptive, ontological classification, sequence, protein structure, publication, taxonomic distribution, and bioinformatics data/model information. By providing an ontology-centered interface, the CARD offers a clearinghouse of information on antibiotic resistance genes, mechanisms, drugs, etc. The left column reflects ontological cross-referencing (see Materials and Methods).
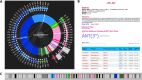
Fig 5
Analysis of the whole genome of Acinetobacter baumannii strain TCDC-AB0715 by the Resistance Gene Identifier (RGI). The A. baumannii strain TCDC-AB0715 is a clinical isolate with resistance to carbapenems, fluoroquinolones, and cephalosporins (26). (A) “Resistance wheel” for A. baumannii strain TCDC-AB0715, predicting resistance to a broad range of antibiotic classes. (B) Details screen of orf0_267, illustrating detection of the aminoglycoside nucleotidyltransferase ANT(3″). (C) Open reading frame (ORF) map of a region of the A. baumannii strain TCDC-AB0715 chromosome, with prediction of β-lactamases TEM-1 and TEM-33 (light blue), aminoglycoside phosphotransferases, nucleotidyltransferases, and acetyltransferases (pink), chloramphenicol acetyltransferase (bright green), sulfonamide-resistant dihydropteroate synthase sul1 (dark green), tetracycline efflux pump tetB (dark pink), and genes implicated in general efflux (dark blue). ORFs unrelated to antibiotic resistance are presented in gray, while non-protein-coding regions are presented in black. The resistance genes identified are consistent with the reported resistance phenotype (26).
Similar articles
- CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database.
Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, Lago BA, Dave BM, Pereira S, Sharma AN, Doshi S, Courtot M, Lo R, Williams LE, Frye JG, Elsayegh T, Sardar D, Westman EL, Pawlowski AC, Johnson TA, Brinkman FS, Wright GD, McArthur AG. Jia B, et al. Nucleic Acids Res. 2017 Jan 4;45(D1):D566-D573. doi: 10.1093/nar/gkw1004. Epub 2016 Oct 26. Nucleic Acids Res. 2017. PMID: 27789705 Free PMC article. - ARDB--Antibiotic Resistance Genes Database.
Liu B, Pop M. Liu B, et al. Nucleic Acids Res. 2009 Jan;37(Database issue):D443-7. doi: 10.1093/nar/gkn656. Epub 2008 Oct 2. Nucleic Acids Res. 2009. PMID: 18832362 Free PMC article. - CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database.
Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. Alcock BP, et al. Nucleic Acids Res. 2020 Jan 8;48(D1):D517-D525. doi: 10.1093/nar/gkz935. Nucleic Acids Res. 2020. PMID: 31665441 Free PMC article. - An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review. - [Overview of antibiotic resistance genes database].
Yang B, Liang J, Liu L, Li X, Wang Q, Ren Y. Yang B, et al. Sheng Wu Gong Cheng Xue Bao. 2020 Dec 25;36(12):2582-2597. doi: 10.13345/j.cjb.200375. Sheng Wu Gong Cheng Xue Bao. 2020. PMID: 33398956 Review. Chinese.
Cited by
- Detection and Genomic Characterisation of Clostridioides difficile from Spinach Fields.
Marcos P, Whyte P, Burgess C, Ekhlas D, Bolton D. Marcos P, et al. Pathogens. 2022 Nov 8;11(11):1310. doi: 10.3390/pathogens11111310. Pathogens. 2022. PMID: 36365061 Free PMC article. - Predicting fungal secondary metabolite activity from biosynthetic gene cluster data using machine learning.
Riedling O, Walker AS, Rokas A. Riedling O, et al. Microbiol Spectr. 2024 Feb 6;12(2):e0340023. doi: 10.1128/spectrum.03400-23. Epub 2024 Jan 9. Microbiol Spectr. 2024. PMID: 38193680 Free PMC article. - Outer membrane vesicles secreted from Actinobacillus pleuropneumoniae isolate disseminating the floR resistance gene to Enterobacteriaceae.
Xu M, Ke H, Zang Y, Gou H, Yang D, Shi K, Zhang K, Li Y, Jiang Z, Chu P, Zhai S, Li C. Xu M, et al. Front Microbiol. 2024 Sep 5;15:1467847. doi: 10.3389/fmicb.2024.1467847. eCollection 2024. Front Microbiol. 2024. PMID: 39301187 Free PMC article. - Draft Genome Sequence of Mycobacterium heraklionense Strain Davo.
Greninger AL, Cunningham G, Chiu CY, Miller S. Greninger AL, et al. Genome Announc. 2015 Jul 23;3(4):e00807-15. doi: 10.1128/genomeA.00807-15. Genome Announc. 2015. PMID: 26205863 Free PMC article. - Genetic and Phenotypic Virulence Potential of Non-O1/Non-O139 Vibrio cholerae Isolated from German Retail Seafood.
Zhang Q, Alter T, Strauch E, Hammerl JA, Schwartz K, Borowiak M, Deneke C, Fleischmann S. Zhang Q, et al. Microorganisms. 2023 Nov 11;11(11):2751. doi: 10.3390/microorganisms11112751. Microorganisms. 2023. PMID: 38004762 Free PMC article.
References
- D'Costa VM, King CE, Kalan L, Morar M, Sung WWL, Schwarz C, Froese D, Zazula G, Calmels F, Debruyne R, Golding GB, Poinar HN, Wright GD. 2011. Antibiotic resistance is ancient. Nature 477:457–461 - PubMed
- Wright GD. 2010. The antibiotic resistome. Expert Opin. Drug Discov. 5:779–788 - PubMed
- Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, Walsh TR. 2009. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53:5046–5054 - PMC - PubMed
- Köser CU, Ellington MJ, Cartwright EJP, Gillespie SH, Brown NM, Farrington M, Holden MTG, Dougan G, Bentley SD, Parkhill J, Peacock SJ. 2012. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog. 8:e1002824.10.1371/journal.ppat.1002824 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical