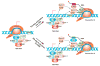SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase - PubMed (original) (raw)
SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase
Xiong Ji et al. Cell. 2013.
Abstract
RNAP II is frequently paused near gene promoters in mammals, and its transition to productive elongation requires active recruitment of P-TEFb, a cyclin-dependent kinase for RNAP II and other key transcription elongation factors. A fraction of P-TEFb is sequestered in an inhibitory complex containing the 7SK noncoding RNA, but it has been unclear how P-TEFb is switched from the 7SK complex to RNAP II during transcription activation. We report that SRSF2 (also known as SC35, an SR-splicing factor) is part of the 7SK complex assembled at gene promoters and plays a direct role in transcription pause release. We demonstrate RNA-dependent, coordinated release of SRSF2 and P-TEFb from the 7SK complex and transcription activation via SRSF2 binding to promoter-associated nascent RNA. These findings reveal an unanticipated SR protein function, a role for promoter-proximal nascent RNA in gene activation, and an analogous mechanism to HIV Tat/TAR for activating cellular genes.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
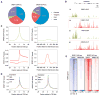
Figure 1. SR Proteins SRSF1 and SRSF2 Interact with DNA on Gene Promoters and RNA on Exonic Regions
(A) Genomic distribution of SR protein ChIP tags (SRSF1 total tags= 10,529,663; SRSF2 total tags = 5,199,318), showing that SRSF1 and SRSF2 have similar binding patterns with a significant fraction mapped to gene promoters in each case. (B) SR protein ChIP-seq and CLIP-seq signals on the representative hnRNPH1 gene. y axis indicates normalized tags per million, with the floor set to 0. The SR CLIP-seq data sets (SRSF1 total tags = 3,694,535; SRSF2 total tags = 4,874,935) on the same MEFs are from the published work (Pandit et al., 2013). (C) Metagene analysis of SRSF2 ChIP-seq (green) and CLIP-seq (red) data at the TSS (based on 23,158 annotated TSS), compared to SRSF2 signals on internal exons (based on 149,352 annotated mouse exons). y axis indicates tags per million per gene. (D) Correlation between SR ChIP-seq signals at the TSS and gene expression analyzed by using all genes with unique and nonoverlapping TSSs. Genes were divided into three groups based on RNA-seq: high (n = 2,829), medium (n = 2,829), and low (n = 2,828). p value is < 2.2 × 10−16 on all pairwise comparisons according to two-tailed Kolmogorov-Smirnov test. y axis indicates tag density per million per gene. (E) Heatmaps of SR-DNA interactions near the TSS in cells depleted of a different SR protein. Raw tag counts from the same amounts of starting cells were used for comparisons (SRSF1 ChIP-seq tags in WT MEFs = 4,538,963; SRSF1 ChIP-seq tags in SRSF2-depleted MEFs = 551,590; SRSF2 ChIP-seq tags in WT MEFs = 9,489,245; SRSF1 ChIP-seq tags in SRSF2-depleted MEFs = 551,933). See also Figure S1.
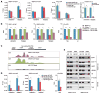
Figure 2. SR Proteins Are Required for RNAP II Pause Release
(A and B) UCSC genome browser views of RNAP II ChIP-seq (detected by N20) and GRO-seq signals on the representative hnRNPH1 gene before and after Dox-induced depletion of SRSF1 (A) or SRSF2 (B) in MEFs. y axis indicates normalized tags per million, with the floor set to 0. (C) Metagene analysis of RNAP II ChIP-seq (top) or GRO-seq (bottom) signals at the TSS (n = 23,037) in response to SRSF2 depletion. SRSF2-bound and unbound genes were separately compared. The differences are significant (p < 2.2 × 10−16) based on two-tailed KS test. y axis indicates normalized tags per million per TSS. (D) Shift of traveling ratio (TR) based on RNAP II ChIP-seq (top) or GRO-seq (bottom) data sets of active genes in response to SRSF2 depletion (n = 5,703, p < 2.2 × 10−16) according to two-tailed KS test in both cases. (E) TR differences based on RNAP II ChIP-seq signals in MEFs depleted of hnRNP A (top) or hnRNP B (bottom). The knockdown effects were verified by western blotting (insets). (F) TR shifts based on RNAP II ChIP-seq (top) or GRO-seq (bottom) correlated with induced gene expression in response to SRSF2 depletion. Averaged changes in gene expression (FDR < 0.05) detected by RNA-seq were plotted against three groups of genes evenly divided according to their TR differences from large to small. See also Figure S2.
Figure 3. Noncoding 7SK RNA Mediates SR Protein Binding to Gene Promoters
(A) ChIP-qPCR analysis of SR protein interaction with two gene promoters (HNRNPH1 and TMSB4X) in MEFs mock treated with DMSO or treated with α-amanitin (left two panels). The effects of α-amanitin on RNAP II occupancy and nascent RNA (produced during nuclear run-on) at the TSS regions of the two genes were determined by ChIP or RT-qPCR (right two panels). (B) ChIP-qPCR analysis of SR protein interaction with gene promoters using cell lysate treated with RNase T1 or RNase H plus anti-7SK oligo (7SK AS) (left two panels). 7SK level was measured by RT-qPCR; U1 snRNA served as a negative control (right). (C) SR protein CLIP-seq signals on the 7SK RNA. IgG CLIP served as a negative control. y axis indicates normalized tags per million, with the floor set to 0. (D) ChIP-qPCR analysis of SR protein interaction with gene promoters in response to degradation of the 7SK RNA by an anti-7SK oligo in MEFs. A scrambled oligo served as a negative control. The far-right panel shows the level of the 7SK RNA measured by RT-qPCR under each treatment condition. (E) Co-IP/western blotting analysis, showing SR proteins as part of the 7SK complex. Data are shown in (A), (B), and (D) as mean ± SD. *p < 0.05 and **p < 0.005 based on Student's t test. See also Figure S3.
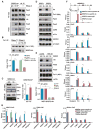
Figure 4. SR Proteins Mediate P-TEFb Release from the 7SK Complex in an RNA-Dependent Manner
(A) Experimental strategy used to fractionate MEFs. Both active and inhibitory components of the 7SK complex are equally distributed between the soluble (S1) and chromatin-bound fraction (P1 or S2). Histone H3 and α-tubulin served as chromatin-bound and unbound markers. (B) ChIP-qPCR analysis of CKD9 and HEXIM1 interactions on four gene promoters in glutaraldehyde-crosslinked MEFs. “Intergenic” indicates a region ∼5 kb upstream the Vim gene promoter. (C) Genome-wide analysis of CDK9 (blue) and HEXIM1 (red) interactions near the TSS(n = 23,037) in glutaraldehyde-crosslinked MEFs. Note some background enrichment with IgG control (green) under this experimental condition. p < 2.2 × 10−16 is calculated based on two-tailed KS test. y axis indicates normalized tags per million per gene. (D) Venn diagrams of genomic interactions between CDK9 and SRSF2 detected by ChIP-seq (left) and the induction of RNAP II pausing on P-TEFb-dependent versus SRSF2-dependent genes (right), indicating extensive physical and functional relationships (p < 2.2 × 10−16, hypergometric test) between these two factors. (E) RNA-dependent release of SR proteins and CDK9 from anti-HEXIM1 IPed 7SK complex. (Top) The strategy for the RNA-mediated P-TEFb release assay. (Bottom) Western blotting analysis of SR proteins and CDK9 released from the 7SK complex with increasing amounts of RNA. Blue and red boxes, respectively, highlight dosage-dependent P-TEFb release induced by the purine-rich ESE and the 2′-O-methyl oligo complementary to the mapped SR-binding site in the 7SK RNA. Data in (A) and (B) are shown as mean ± SD. See also Figure S4.
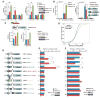
Figure 5. 7SK RNA Connects SR Proteins to RNAP II, and SR Proteins Are Required for SEC Recruitment to Gene Promoters
(A) Co-IP/western blotting analysis, showing RNA-dependent association of RNAP II with HA-tagged SRSF2. (B) Reciprocal co-IP/western blotting analysis, demonstrating RNA-dependent association of HA-tagged SRSF2 with IPed RNAP II. Levels of the 7SK RNA were determined by RT-qPCR under different treatment conditions (bottom). (C) Ribo-IP analysis, showing slightly increased association of CDK9 with 7SK in SRSF2-depleted MEFs. (Right) Levels of CDK9-associated 7SK quantified by RT-qPCR. (D) Western blotting analysis, showing diminished Ser2-phosphorylated RNAP II (Pser2) in SR protein-depleted MEFs. Specific antibodies were used to detect total RNAP II (N20), Ser2- and Ser5-phosphorylated RNAP II (Pser2 and Pser5), CDK9, AFF4, and HEXIM1 before and after Dox-induced depletion of SRSF1 or SRSF2. α-tubulin served as loading control. (E) Restoration of RNAP II Ser2 phosphorylation in MEFs depleted of both SRSF2 and 7SK RNA. (Bottom) Levels of 7SK determined by RT-qPCR. (F) Requirement of SRSF2 for efficient recruitment of the superelongation complex (SEC) to gene promoters. In response to SRSF2 depletion, Ser2-phosphorylated RNAP II and two common components (CDK9 and AFF4) of SEC were dramatically reduced on HnRNPH1 gene, but the levels of HEXIM1 and chromatin-bound Brd4 were unaffected. (G) Brd4 requirement for the recruitment of CDK9 and SRSF2 to gene promoters. Data in (B), (C), (E), (F), and (G) are shown as mean ± SD. *p < 0.05 and **p < 0.005 based on Student's t test. See also Figure S5.
Figure 6. High-Affinity RNA Elements Mediate Transcriptional Activation by SRSF2
(A) Luciferase assay in transfected HEK293T cells, showing activation of the HIV-1 promoter by overexpressed V5-tagged SRSF1 or HA-tagged SRSF2 (inset). SRSF1 activated the HSV-driven reporter at the translational level (no induction of mRNA), but SRSF2 had no effect on HSV. None of the SR proteins activated the CMV promoter. (B) Luciferase assay of reporters constructed from the hnRNPH1 gene in response to SR protein overexpression (left) or RNAi (right). Specific constructs are illustrated on the right. (C) Tethered SRSF2 activated transcription from an HSV-based reporter containing a specific PUF binding motif (red box). The SRSF2-PUF fusion protein activated transcription, whereas SRSF1-PUF fusion protein stimulated translation of the reporter. A plasmid not carrying any SR-coding sequences served as a negative control. (D–F) Schematic presentation of HSV-based reporters (D). Dual luciferase assays based on PCMV (internal control) and HSV-ESE reporters in response to SRSF2 overexpression (E) or RNAi (F). (Insets) Protein levels monitored by western blotting. (G) Comparison of traveling ratio (TR) of transcriptionally engaged RNAP II (based on GRO-seq signals) on two groups of genes with high (blue) or low/no (green) SRSF2 CLIP-seq signals on their 300 nt TSS-associated RNA. Genes with lower TR are more linked than those with higher TR to SRSF2 binding on RNA near the TSS. The differences are highly significant (p < 2.2 × 10−16) based on the two-tailed KS test. Data in (A), (B), (C), (E), and (F) are shown as mean ± SD. *p < 0.05 and **p < 0.005 based on Student's t test. See also Figure S6.
Figure 7. A Working Model for SR Protein-Dependent Transcriptional Activation
SR proteins and the 7SK RNA complex are intimately associated with genomic DNA near the promoter-proximal region (left). This proximity may allow more efficient local switches during gene activation. During Tat-dependent activation of the HIV-1 promoter (upper-right), Tat binding to TAR induces relocation of P-TEFb (CDK9:cyclin T) from the 7SK complex to paused RNAP II. This process may be facilitated by direct protein-protein interactions between Tat and cyclin T and between Brd4 and CDK9. It is currently unclear whether released P-TEFb is directly recruited to RNAP II or indirectly via the nucleoplasmic pool, as indicated by the dashed arrows. During transcription pause release on cellular genes (lower-right), SR proteins are also associated with gene promoters as part of the 7SK complex. We speculate that, by taking advantage of local assembly, an SRSF2-binding site (ESE) emerging from RNAP II may induce the SR protein to switch from the 7SK RNA to nascent RNA, thereby triggering the coordinated release of P-TEFb from the 7SK complex. Again, the released P-TEFb may go through two separate routes before being recruited to paused RNAP II at the TSS, as indicated by the dashed arrows. In both the Tat/TAR and SR/ESE systems, chromatin-bound Brd4 may enhance the association of released P-TEFb with RNAP II at the TSS. Recruited P-TEFb will phosphorylate RNAP II and some key factors, such as NELF and DSIF, resulting in transcription pause release.
Similar articles
- Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein.
Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. Bartholomeeusen K, et al. J Biol Chem. 2012 Oct 19;287(43):36609-16. doi: 10.1074/jbc.M112.410746. Epub 2012 Sep 5. J Biol Chem. 2012. PMID: 22952229 Free PMC article. - KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II.
McNamara RP, Reeder JE, McMillan EA, Bacon CW, McCann JL, D'Orso I. McNamara RP, et al. Mol Cell. 2016 Jan 7;61(1):39-53. doi: 10.1016/j.molcel.2015.11.004. Epub 2015 Dec 24. Mol Cell. 2016. PMID: 26725010 Free PMC article. - The 7SK/P-TEFb snRNP controls ultraviolet radiation-induced transcriptional reprogramming.
Studniarek C, Tellier M, Martin PGP, Murphy S, Kiss T, Egloff S. Studniarek C, et al. Cell Rep. 2021 Apr 13;35(2):108965. doi: 10.1016/j.celrep.2021.108965. Cell Rep. 2021. PMID: 33852864 - Cracking the control of RNA polymerase II elongation by 7SK snRNP and P-TEFb.
C Quaresma AJ, Bugai A, Barboric M. C Quaresma AJ, et al. Nucleic Acids Res. 2016 Sep 19;44(16):7527-39. doi: 10.1093/nar/gkw585. Epub 2016 Jul 1. Nucleic Acids Res. 2016. PMID: 27369380 Free PMC article. Review. - Transcription elongation control by the 7SK snRNP complex: Releasing the pause.
McNamara RP, Bacon CW, D'Orso I. McNamara RP, et al. Cell Cycle. 2016 Aug 17;15(16):2115-2123. doi: 10.1080/15384101.2016.1181241. Epub 2016 May 6. Cell Cycle. 2016. PMID: 27152730 Free PMC article. Review.
Cited by
- A large-scale binding and functional map of human RNA-binding proteins.
Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Xiao R, Blue SM, Chen JY, Cody NAL, Dominguez D, Olson S, Sundararaman B, Zhan L, Bazile C, Bouvrette LPB, Bergalet J, Duff MO, Garcia KE, Gelboin-Burkhart C, Hochman M, Lambert NJ, Li H, McGurk MP, Nguyen TB, Palden T, Rabano I, Sathe S, Stanton R, Su A, Wang R, Yee BA, Zhou B, Louie AL, Aigner S, Fu XD, Lécuyer E, Burge CB, Graveley BR, Yeo GW. Van Nostrand EL, et al. Nature. 2020 Jul;583(7818):711-719. doi: 10.1038/s41586-020-2077-3. Epub 2020 Jul 29. Nature. 2020. PMID: 32728246 Free PMC article. - RNA splicing factors as oncoproteins and tumour suppressors.
Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. Dvinge H, et al. Nat Rev Cancer. 2016 Jul;16(7):413-30. doi: 10.1038/nrc.2016.51. Epub 2016 Jun 10. Nat Rev Cancer. 2016. PMID: 27282250 Free PMC article. Review. - Alternative splicing regulates the expression of G9A and SUV39H2 methyltransferases, and dramatically changes SUV39H2 functions.
Mauger O, Klinck R, Chabot B, Muchardt C, Allemand E, Batsché E. Mauger O, et al. Nucleic Acids Res. 2015 Feb 18;43(3):1869-82. doi: 10.1093/nar/gkv013. Epub 2015 Jan 20. Nucleic Acids Res. 2015. PMID: 25605796 Free PMC article. - Analog-sensitive cell line identifies cellular substrates of CDK9.
Decker TM, Forné I, Straub T, Elsaman H, Ma G, Shah N, Imhof A, Eick D. Decker TM, et al. Oncotarget. 2019 Dec 10;10(65):6934-6943. doi: 10.18632/oncotarget.27334. eCollection 2019 Dec 10. Oncotarget. 2019. PMID: 31857848 Free PMC article. - Targeting Processive Transcription Elongation via SEC Disruption for MYC-Induced Cancer Therapy.
Liang K, Smith ER, Aoi Y, Stoltz KL, Katagi H, Woodfin AR, Rendleman EJ, Marshall SA, Murray DC, Wang L, Ozark PA, Mishra RK, Hashizume R, Schiltz GE, Shilatifard A. Liang K, et al. Cell. 2018 Oct 18;175(3):766-779.e17. doi: 10.1016/j.cell.2018.09.027. Cell. 2018. PMID: 30340042 Free PMC article.
References
- Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol. 2005;17:251–256. - PubMed
- Berkhout B, Gatignol A, Rabson AB, Jeang KT. TAR-independent activation of the HIV-1 LTR: evidence that tat requires specific regions of the promoter. Cell. 1990;62:757–767. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- HG004659/HG/NHGRI NIH HHS/United States
- R01 GM052872/GM/NIGMS NIH HHS/United States
- R01 HG004659/HG/NHGRI NIH HHS/United States
- GM052872/GM/NIGMS NIH HHS/United States
- GM049369/GM/NIGMS NIH HHS/United States
- R01 GM049369/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials