A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila - PubMed (original) (raw)
A genome-wide RNAi screen draws a genetic framework for transposon control and primary piRNA biogenesis in Drosophila
Felix Muerdter et al. Mol Cell. 2013.
Abstract
A large fraction of our genome consists of mobile genetic elements. Governing transposons in germ cells is critically important, and failure to do so compromises genome integrity, leading to sterility. In animals, the piRNA pathway is the key to transposon constraint, yet the precise molecular details of how piRNAs are formed and how the pathway represses mobile elements remain poorly understood. In an effort to identify general requirements for transposon control and components of the piRNA pathway, we carried out a genome-wide RNAi screen in Drosophila ovarian somatic sheet cells. We identified and validated 87 genes necessary for transposon silencing. Among these were several piRNA biogenesis factors. We also found CG3893 (asterix) to be essential for transposon silencing, most likely by contributing to the effector step of transcriptional repression. Asterix loss leads to decreases in H3K9me3 marks on certain transposons but has no effect on piRNA levels.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
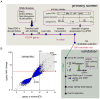
Figure 1. A genome-wide RNAi screen for piRNA pathway components acting in the somatic compartment of Drosophila ovaries
A) A workflow of the primary RNAi screen in ovarian somatic sheet cells (OSS) and validation of primary hit candidates in vivo is shown. Each gene in the Drosophila genome was knocked down with one or more dsRNAs. 5 days after transfection, cells were tested for increased levels of the gypsy retrotransposon. The primers and the hydrolysis probe used for the qPCR are shown (FP: forward primer, P: hydrolysis probe, RP: reverse primer). The dashed line indicates the ~5kb segment not present in the subgenomic transcript. 288 genes were further tested in vivo using the Gal4/UAS system to drive hairpin RNAs (hpRNAs) within the traffic jam (TJ) expression domain. B) All transfected wells were assayed for levels of gypsy and one reference gene for normalization. Levels of gypsy expression are displayed as z-scores and fold change. The cutoffs for both z-score (<−1.9) and fold change (>3) are indicated as red lines. The shaded area shows the selection of primary hit candidates. Three positive controls (piwi, armi, zuc) and one negative control (white) are marked as red dots. Only wells that passed the filter for primary datapoint selection are shown. For all primary datapoints see Table S1. See also Figure S1.
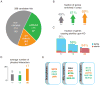
Figure 2. Primary candidates were validated in vivo
A) The number of hit candidates that validated (va) or did not validate (nv) in vivo is shown. Genes that upon knockdown caused severe developmental defects and therefore could not be assayed are also indicated (dd). A full list of validated fly lines and corresponding transposon derepression information is available in Table S3. B) Validated hits are preferentially expressed in ovaries. The percentage of genes that are enriched in ovaries compared to whole fly is shown for the three categories. This data is based on mRNA signals on Affymetrix expression arrays available from FlyAtlas (Chintapalli et al., 2007). C) The fraction of genes causing sterility upon knockdown is shown. Each small box represents one gene, with blue and red indicating if flies were fertile or sterile upon knockdown. D) The degree to which a gene may represent a node in a network in each of the classifications was measured by number of physical interactors. Interaction data from BIOGRID was used for this analysis. E) Components of the Drosophila sumoylation pathway, the Nonspecific Lethal Complex, and proteins involved in nuclear export are primary hits that validate in vivo. WAH could not be validated in vivo because no RNAi fly was available at the time of submission (red asterisk). The text coloring of each gene indicates the result of the validation screen and is consistent with the categories in panel A. See also Figure S2.
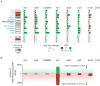
Figure 3. RNA-seq shows changes in gene and transposon expression upon knockdown of top candidates in vivo
A) A subset of somatically expressed transposons is derepressed in the indicated KD. The classification of transposons according to Malone et al. (2009), is indicated in orange (germline dominant), grey (intermediate), blue (soma dominant) and black (unclassified). The absolute abundance of reads in control knockdown mapping to each transposon is shown in shades of grey. The log2 fold change of each target gene versus a negative control (aub) is shown. Color of the bars represent the significance of these fold changes and are indicated as an adjusted p-value (FDR). Green indicates highly significant differences (p<=0.05), yellow moderately significant changes (0.05<p<=0.1) and red non-significant changes (0.1<p<=1), based on two biological replicates. Each knockdown is normalized to aub knockdown controls from their corresponding library (GD or KK). For differences in transposon abundance levels between both aub controls, see Figure S3. B) The number of genes differentially expressed (p-adj<0.05) in each knockdown with respect to the control is shown. Green bars indicate the number of genes that have higher expression levels in the knockdown fly line, while red bars designate the number of genes with higher levels in the aub negative control.
Figure 4. Biogenesis of small RNAs from somatic clusters and transposons is affected in knockdowns of a subset of top candidate genes
A) Percentages of total unique mappers (sense species, >23nt) to flamenco in each knockdown (as indicated) in relation to the control knockdown are shown. B) The internal rankings for three representative piRNA clusters based on their representation in piRNA populations are displayed. Expression bias towards either domain (soma or germline) is indicated. Cluster definitions are in concordance with Brennecke et al. (2007). C) The size profiles of piRNAs mapping in sense orientation to flamenco in each knockdown (as in A) are plotted as total read count per million genomic mappers. As a control, we show that levels of microRNAs do not change in knockdowns versus negative control (Figure S4). D) piRNAs mapping to a subset of somatically expressed transposons are reduced when gene expression of a subset of top hits is disrupted. The classification of transposons according to Malone et al. (2009), is indicated in orange (germline dominant), grey (intermediate), blue (soma dominant) and black (unclassified). The absolute abundance of antisense piRNAs in an aub control mapping to each transposon is shown in shades of grey. The log2 fold change of each target gene versus a negative control is shown. See also Figure S4.
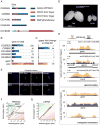
Figure 5. Disruption of CG3893 function has a severe impact on transposon silencing
A) The five members of the Drosophila uncharacterized protein family UPF0224 and their domain structures are diagrammed. The conserved domains are highlighted as colored boxes. B) All five family members are weakly expressed in OSS cells. piwi and ago3 expression levels are shown for comparison. Expression levels are based on the modENCODE cell line expression data and are displayed as reads per kilobase per million mapped reads (rpkm). C) CG3893, but no other members of its protein family, has a strong impact on transposon silencing upon knockdown in OSS cells. Effects of knockdown of ago3 and piwi are shown for comparison. Numbers represent fold changes of gypsy levels in respect to the median fold change of the corresponding plate in the primary screen. D) The ovarian morphology of flies heterozygous or homozygous for a P-element insertion in CG3893 is shown (204406, Kyoto Drosophila Genetic Resource Center). For a more detailed view of the insertion and expression levels see Figure S5. E) Tagged CG3893 co-localizes with Piwi in the nucleus of OSS cells when overexpressed in transient transfections. Nuclear Hoechst staining is blue, GFP tagged CG3893 is green and RFP tagged Piwi or ΔNT-Piwi is shown in red (Saito et al., 2009). F) Transposons are highly upregulated upon disruption of CG3893 in the P-element insertion line. A scatter plot of reads per million (rpm) is shown for RNA-seq of heterozygous versus homozygous flies. Each dot represents one transposon consensus sequence. Only sequences mapping in the sense orientation are taken into account. G) piRNA levels are not affected by CG3893 disruption. The number of piRNA reads mapped to the same transposon consensus sequences as in F) is expressed in reads per million. H) Levels of H3K9me3 decrease dramatically on a subset of transposons upon depletion of CG3893. Density plots for normalized H3K9me3 ChIP-seq reads over three transposons, gtwin, gypsy and Het-A are shown. Yellow distributions correspond to levels in heterozygous flies and blue distributions to the homozygous state. The upper box shows three distinct genomic peaks over transposon insertions, the lower box shows the corresponding consensus sequences. For all identified H3K9me3 peaks and their read densities see Figure S5C. See also Figure S5.
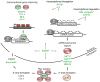
Figure 6. Potential roles for newly identified piRNA pathway components
Known piRNA components are shown as bubbles. The newly identified genes are shown in bold text colored according to their validation status (color code as in figure 2; Pol2, RNA polymerase 2; red hexagons represent H3K9me3). Red asterisks denote genes that validated in the germline screen done in parallel (Czech et al., submitted).
Similar articles
- A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway.
Czech B, Preall JB, McGinn J, Hannon GJ. Czech B, et al. Mol Cell. 2013 Jun 6;50(5):749-61. doi: 10.1016/j.molcel.2013.04.007. Epub 2013 May 9. Mol Cell. 2013. PMID: 23665227 Free PMC article. - Maternally inherited piRNAs direct transient heterochromatin formation at active transposons during early Drosophila embryogenesis.
Fabry MH, Falconio FA, Joud F, Lythgoe EK, Czech B, Hannon GJ. Fabry MH, et al. Elife. 2021 Jul 8;10:e68573. doi: 10.7554/eLife.68573. Elife. 2021. PMID: 34236313 Free PMC article. - Multiple roles for Piwi in silencing Drosophila transposons.
Rozhkov NV, Hammell M, Hannon GJ. Rozhkov NV, et al. Genes Dev. 2013 Feb 15;27(4):400-12. doi: 10.1101/gad.209767.112. Epub 2013 Feb 7. Genes Dev. 2013. PMID: 23392609 Free PMC article. - piRNA-Guided Transposon Silencing and Response to Stress in Drosophila Germline.
Ho S, Theurkauf W, Rice N. Ho S, et al. Viruses. 2024 Apr 30;16(5):714. doi: 10.3390/v16050714. Viruses. 2024. PMID: 38793595 Free PMC article. Review. - The piRNA pathway in Drosophila ovarian germ and somatic cells.
Sato K, Siomi MC. Sato K, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review.
Cited by
- PIWI-interacting RNAs: from generation to transgenerational epigenetics.
Luteijn MJ, Ketting RF. Luteijn MJ, et al. Nat Rev Genet. 2013 Aug;14(8):523-34. doi: 10.1038/nrg3495. Epub 2013 Jun 25. Nat Rev Genet. 2013. PMID: 23797853 Review. - "Dot COM", a nuclear transit center for the primary piRNA pathway in Drosophila.
Dennis C, Zanni V, Brasset E, Eymery A, Zhang L, Mteirek R, Jensen S, Rong YS, Vaury C. Dennis C, et al. PLoS One. 2013 Sep 9;8(9):e72752. doi: 10.1371/journal.pone.0072752. eCollection 2013. PLoS One. 2013. PMID: 24039799 Free PMC article. - Rapid evolution and conserved function of the piRNA pathway.
Parhad SS, Theurkauf WE. Parhad SS, et al. Open Biol. 2019 Jan 31;9(1):180181. doi: 10.1098/rsob.180181. Open Biol. 2019. PMID: 30958115 Free PMC article. Review. - Daedalus and Gasz recruit Armitage to mitochondria, bringing piRNA precursors to the biogenesis machinery.
Munafò M, Manelli V, Falconio FA, Sawle A, Kneuss E, Eastwood EL, Seah JWE, Czech B, Hannon GJ. Munafò M, et al. Genes Dev. 2019 Jul 1;33(13-14):844-856. doi: 10.1101/gad.325662.119. Epub 2019 May 23. Genes Dev. 2019. PMID: 31123065 Free PMC article. - Chromatin-associated RNAs as facilitators of functional genomic interactions.
Li X, Fu XD. Li X, et al. Nat Rev Genet. 2019 Sep;20(9):503-519. doi: 10.1038/s41576-019-0135-1. Nat Rev Genet. 2019. PMID: 31160792 Free PMC article. Review.
References
- Andrulis ED, Werner J, Nazarian A, Erdjument-Bromage H, Tempst P, Lis JT. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature. 2002;420:837–841. - PubMed
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete Small RNA-Generating Loci as Master Regulators of Transposon Activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
- Bucheton A. The relationship between the flamenco gene and gypsy in Drosophila: how to tame a retrovirus. Trends in genetics: TIG. 1995;11:349–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5R01GM062534/GM/NIGMS NIH HHS/United States
- T32 GM065094/GM/NIGMS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R01 GM067761/GM/NIGMS NIH HHS/United States
- R01 GM062534/GM/NIGMS NIH HHS/United States
- R37 GM062534/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials