The genetic makeup of the Drosophila piRNA pathway - PubMed (original) (raw)
The genetic makeup of the Drosophila piRNA pathway
Dominik Handler et al. Mol Cell. 2013.
Abstract
The piRNA (PIWI-interacting RNA) pathway is a small RNA silencing system that acts in animal gonads and protects the genome against the deleterious influence of transposons. A major bottleneck in the field is the lack of comprehensive knowledge of the factors and molecular processes that constitute this pathway. We conducted an RNAi screen in Drosophila and identified ~50 genes that strongly impact the ovarian somatic piRNA pathway. Many identified genes fall into functional categories that indicate essential roles for mitochondrial metabolism, RNA export, the nuclear pore, transcription elongation, and chromatin regulation in the pathway. Follow-up studies on two factors demonstrate that components acting at distinct hierarchical levels of the pathway were identified. Finally, we define CG2183/Gasz as an essential primary piRNA biogenesis factor in somatic and germline cells. Based on the similarities between insect and vertebrate piRNA pathways, our results have far-reaching implications for the understanding of this conserved genome defense system.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
Figure 1
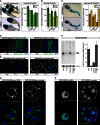
An RNAi Screen for Somatic piRNA Pathway Factors (A) Cartoon of a Drosophila ovariole with somatic cells in green and germline cells in beige. (B) Schematic representations of the Drosophila germline and somatic piRNA pathways focusing on the three PIWI family proteins and the biogenesis routes of their bound piRNAs. (C) Detailed model of the somatic piRNA pathway. Known pathway members are placed at their functional positions based on literature (TGS, transcriptional gene silencing; PTGS, posttranscriptional gene silencing). (D) Illustration of the _gypsy_-lacZ reporter. Shown are normalized profiles of ovarian and OSC piRNAs (sense up, antisense down) mapping to the gypsy TE and the gypsy sequence portion present in the reporter. (E) Shown are β-gal stainings of ovarioles as readout for the _gypsy_-lacZ reporter from flamenco restrictive (upper panel) or flamenco permissive flies (lower panel). (F) Bar chart showing expression levels (average RPKM values (log10 scale) obtained from two independent RNA-seq experiments) of all annotated Drosophila genes in OSCs. Several known somatic piRNA pathway factors (violet) and germline-specific control genes (green) are indicated. (G) Illustration of the screen workflow. Indicated are the numbers of tested RNAi lines and the corresponding number of genes for the primary and secondary screens. (H) Shown are β-gal stainings of representative egg chambers indicating major staining categories used for the evaluation of the screen crosses (left images, wild-type morphology; right image, distorted morphology).
Figure 2
High Sensitivity, Specificity, and Reproducibility of the Screen (A) Indicated to the left is the impact on ovarian morphology observed upon knockdown of the 83 genes encoding for ribosomal proteins. The two bar charts are based on this analysis and indicate the percentage of effective VDRC lines (left) and the corresponding false negative rate (8%) at the gene level if approximately two lines per gene were tested (right). (B) Bar chart illustrating the ovarian morphology phenotype observed for all genes tested in the screen. At least one line per gene had to fall into the indicated categories. Also shown are the most enriched gene ontology (GO) categories (p values corrected for multiple testing) for the set of 663 genes that classified for the “no ovary” phenotype. (C) Indicated are the percentages of genes flagged with the “no ovary” or “distorted morphology” phenotypes when all tested genes were split into ten bins according to their expression level (gray triangle). Bins 1–10 are equally sized bins (n = 682) of all expressed genes (RPKM > 1), while bin 0 contains 334 randomly tested genes expressed below RPKM = 1. (D) Shown are the _gypsy_-lacZ staining results from the screen for available VDRC lines targeting the eight known piRNA pathway factors. (E) Bar chart summarizing the _gypsy_-lacZ staining results ranging from weak to strong for all genes tested in the screen. (F) Indicated are the percentages of genes scoring with the indicated _gypsy_-lacZ intensities when all tested genes were split into ten bins according to their expression level (gray triangle). Bins 1–10 are equally sized bins (n = 682) of all expressed genes (RPKM > 1), while bin 0 contains 334 randomly tested genes expressed below RPKM = 1. (G) Compared are _gypsy_-lacZ intensities as well as the morphology phenotypes for 30 screen-positive genes tested with shRNA lines or VDRC lines (NA, not analyzable due to a “no ovary” phenotype). (H) Shown are box plots displaying the fold changes in steady-state RNA levels (based on RT-qPCR) of lacZ, ZAM, and gypsy normalized to control knockdowns. Tested were all RNAi lines (numbers given at the bottom) falling into the five staining categories (color coded) and 11 control lines (gray). Box plots show median (line), 25th–75th percentile (box) ± 1.5 interquartile range; circles represent outliers.
Figure 3
Genetic Classification of Germline-Specific, Soma-Specific, and Common piRNA Factors (A) Illustration of the _Burdock_-lacZ reporter. Shown are normalized profiles of ovarian and OSC piRNAs (sense up, antisense down) mapping to the Burdock element and the Burdock sequence portion present in the 3′ UTR of the reporter, which expresses β-gal under control of the germline-specific nanos promoter. (B) Bar diagram displaying the sense/antisense overlap patterns of the ovarian piRNA population mapping to the Burdock element. The red bar at 10 nt indicates a significant ping-pong signature. (C) Bar plot illustrating normalized piRNA levels (%) antisense to the Burdock TE in ovaries from indicated germline-specific knockdowns (MTD × shRNA) compared to control levels. (D) Shown are β-gal stainings of egg chambers expressing the _Burdock_-lacZ reporter and germline knockdowns (KD) for the indicated genes (w[1118] serves as negative control). (E) Listed are all genes known to be specific for the germline piRNA pathway (orange set) and all genes scoring in the somatic screen (intermediate-weak or stronger; no mitochondrial genes). Based on the staining intensities observed with the _Burdock_-lacZ reporter (GL/left columns) or the _gypsy_-lacZ reporter (S/right columns), genes were grouped into germline-specific, common, and soma-specific classes. All genes previously linked to the piRNA pathway are marked as “previously described.” Genes not interpretable in the germline test are shown in the lower left panel. Asterisks indicate genes tested with an shRNA line because no VDRC line was available (AGO3, krimper) or the VDRC line is not functional (SoYb). (F) Box plots showing fold deregulations of HeT-A, Burdock, lacZ, and act5c RNA levels upon knockdown of all screen-scoring VDRC lines. Lines were grouped into three staining categories (blue) based on their effect on the _Burdock_-lacZ reporter. Germline-specific factors (orange) are represented as a separate group.
Figure 4
Key Processes and Factors Involved in the Somatic piRNA Pathway (A) Box plots showing the fold enrichment of gene expression (based on FlyAtlas) in ovaries or larval CNS versus whole flies for all tested genes (gray) and the set of positive screen hits (blue); p values were determined by Wilcoxon signed rank test. (B) Shown are the most significantly enriched GO terms among the 144 scoring genes in the screen (p values corrected for multiple testing). Mitochondria-related terms are in red. (C) Listed are all genes with annotated mitochondrial function and their respective _gypsy_-lacZ staining and ovarian morphology phenotypes observed in the somatic screen. (D) Box plots showing fold increases in lacZ, gypsy, and ZAM steady-state RNA levels for the set of mitochondrial gene knockdowns (green box) compared to nonscoring genes (gray box); p values were determined by Wilcoxon signed rank test. Box plots are defined in Figure 2H. (E) Significantly enriched GO terms among screen hits without mitochondria associated genes (p values corrected for multiple testing). (F) Cartoon depicting functional groups of genes identified in the screen. Factors were grouped and placed into nucleus or cytoplasm based on their annotated functions or by identification of orthologous genes with annotated functions. Genes involved in the various processes are indicated together with the _gypsy_-lacZ staining results.
Figure 5
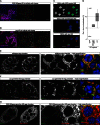
CG2183/Gasz and CG9754 Are Essential for the Somatic piRNA Pathway (A) Left panels show β-gal stainings of ovarioles as readout for _gypsy_-lacZ silencing upon soma knockdown of CG2183/gasz or CG9754. The bar diagram depicts fold increases in RNA levels of indicated TEs in ovaries with soma knockdown of armi, CG2183/gasz, or CG9754 (averages of three biological replicates; error bars, SD; normalized to control knockdown). (B) Displayed are fold increases in RNA levels of indicated TEs in OSCs upon siRNA-mediated knockdown of armi, CG2183/gasz, or CG9754 siRNA in OSCs (averages of three biological replicates; error bars, SD; normalized to control knockdown). (C) Left panels show β-gal stainings of ovarioles as readout for _Burdock_-lacZ silencing upon germline knockdown of CG2183/gasz or CG9754. The bar diagram depicts fold increases in RNA levels of indicated TEs in ovaries with germline knockdown of armi, CG2183/gasz, or CG9754 (averages of three biological replicates; error bars, SD; normalized to control knockdown). (D) Confocal sections (scale bars, 10 μm) through the follicular epithelium of egg chambers stained for Piwi (monochrome panel), DNA (blue), and the clonal marker (green). Cells within the clone (dashed lines mark clone boundaries in the monochrome panels) express dsRNAs against CG2183/gasz (left panel) or CG9754 (right panel). (E) Confocal sections (scale bars, 10 μm) through egg chambers stained for Piwi (monochrome panel and green) and DNA (blue). Knockdown of CG2183/gasz (left) and CG9754 (right) was specifically activated in germline cells. (F) Northern blot analysis of piRNA levels in OSC total RNA upon siRNA-mediated knockdowns of GFP (control), armi, CG9754, or CG2183/gasz. One representative blot probed for idefix piRNA (top) and then reprobed for miR-310 (bottom) is shown. The bar diagram indicates quantified results (normalized to miR-310) based on three independent experiments (error bars, SD.). (G) Confocal sections (scale bars, 10 μm) of OSCs (left panels) or egg chambers (right panels) expressing GFP-CG2183/Gasz stained for DNA (blue). Monochrome panels show the GFP signal separately. (H) Confocal sections (scale bars, 10 μm) of OSCs (left panels) or egg chambers (right panels) expressing GFP-CG9754 stained for DNA (blue). Monochrome panels show the GFP signal separately. See also Figure S1.
Figure 6
CG2183/Gasz Is an Uncharacterized piRNA Biogenesis Factor (A) Confocal sections (scale bars, 10 μm) of OSCs transfected with YFP-CG2183/Gasz (yellow) and Zuc-GFP (green) expression constructs and stained for mitochondria (MitoTracker; red) and DNA (blue). (B) Confocal sections (scale bars, 10 μm) of OSCs transfected with Zuc-GFP and GFP-CG2183/Gasz expression constructs (green) lacking the respective transmembrane domains and stained for mitochondria (MitoTracker; red) and DNA (blue). (C) Confocal sections (scale bars, 10 μm) through egg chambers stained for Piwi, Aub, or AGO3. Control knockdown or CG2183/gasz knockdown was specifically activated in germline cells. (D) Left panels show length profiles of normalized small RNA populations from ovaries with control (upper) or CG2183/gasz (lower) germline knockdown split into miRNAs (small insets) and remaining RNAs (siRNA and piRNA populations indicated). Right panels show respective length profiles of repeat-derived small RNAs only (red, antisense; blue, sense). (E) Normalized piRNA profiles (genome unique; sense up, antisense down) from ovaries with indicated germline knockdowns mapping to cluster 42AB. (F) Scatter plots showing normalized antisense piRNA levels (log2 values) mapping to soma-dominant (green), intermediate (yellow), or germline-dominant (black) TEs from ovaries with indicated germline knockdowns (Pearson correlation [r] based on all TEs). (G) Bar chart displaying the adenosine content at position 10 for sense piRNAs mapping to TEs isolated from ovaries with indicated germline knockdowns. Black lines indicate the expected level based on the average 10A content at positions 2–9 and 11–23. (H) Shown are ping-pong signatures of germline-dominant TEs based on piRNAs from ovaries with indicated germline knockdowns. TEs were ordered according to their ping-pong signature in the VDRC control library. See also Figure S2.
Figure 7
CG2183/Gasz Recruits Armitage to Mitochondria (A and B) Confocal sections (scale bars, 10 μm) through the follicular epithelium of egg chambers stained for Armi (green), Vret (red), and DNA (blue). Knockdown of CG2183/gasz (A) or zuc (B) was clonally induced (clonal marker in magenta; clone borders marked by dashed line). (C) Confocal sections (scale bars, 10 μm) of OSCs with indicated siRNA-mediated knockdowns stained for Piwi (green), Armi (red), and DNA (blue). (D) Quantification of Piwi-Armi colocalization based on (C). The fraction of Piwi-positive pixels colocalizing with Armi-positive pixels is indicated. Box plots are based on six quantified images per knockdown with ∼30 cells each. (E–G) Confocal sections (scale bars, 10 μm) through egg chambers with indicated genotype stained for Armi (green), mitochondria (red), and DNA (blue). Overview panels are shown to the left and high magnification images to the right. Colocalization of Armi and mitochondria in the merge panels appears yellow. See also Figures S3–S5.
Similar articles
- Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila.
Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ. Haase AD, et al. Genes Dev. 2010 Nov 15;24(22):2499-504. doi: 10.1101/gad.1968110. Epub 2010 Oct 21. Genes Dev. 2010. PMID: 20966049 Free PMC article. - A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway.
Czech B, Preall JB, McGinn J, Hannon GJ. Czech B, et al. Mol Cell. 2013 Jun 6;50(5):749-61. doi: 10.1016/j.molcel.2013.04.007. Epub 2013 May 9. Mol Cell. 2013. PMID: 23665227 Free PMC article. - An in vivo RNAi assay identifies major genetic and cellular requirements for primary piRNA biogenesis in Drosophila.
Olivieri D, Sykora MM, Sachidanandam R, Mechtler K, Brennecke J. Olivieri D, et al. EMBO J. 2010 Oct 6;29(19):3301-17. doi: 10.1038/emboj.2010.212. Epub 2010 Sep 3. EMBO J. 2010. PMID: 20818334 Free PMC article. - The piRNA pathway in Drosophila ovarian germ and somatic cells.
Sato K, Siomi MC. Sato K, et al. Proc Jpn Acad Ser B Phys Biol Sci. 2020;96(1):32-42. doi: 10.2183/pjab.96.003. Proc Jpn Acad Ser B Phys Biol Sci. 2020. PMID: 31932527 Free PMC article. Review. - The piRNA pathway: a fly's perspective on the guardian of the genome.
Senti KA, Brennecke J. Senti KA, et al. Trends Genet. 2010 Dec;26(12):499-509. doi: 10.1016/j.tig.2010.08.007. Trends Genet. 2010. PMID: 20934772 Free PMC article. Review.
Cited by
- PIWI-interacting RNAs: from generation to transgenerational epigenetics.
Luteijn MJ, Ketting RF. Luteijn MJ, et al. Nat Rev Genet. 2013 Aug;14(8):523-34. doi: 10.1038/nrg3495. Epub 2013 Jun 25. Nat Rev Genet. 2013. PMID: 23797853 Review. - The Mi-2 nucleosome remodeler and the Rpd3 histone deacetylase are involved in piRNA-guided heterochromatin formation.
Mugat B, Nicot S, Varela-Chavez C, Jourdan C, Sato K, Basyuk E, Juge F, Siomi MC, Pélisson A, Chambeyron S. Mugat B, et al. Nat Commun. 2020 Jun 4;11(1):2818. doi: 10.1038/s41467-020-16635-5. Nat Commun. 2020. PMID: 32499524 Free PMC article. - Escalation of genome defense capacity enables control of an expanding meiotic driver.
Chen P, Pan KC, Park EH, Luo Y, Lee YCG, Aravin AA. Chen P, et al. Proc Natl Acad Sci U S A. 2025 Jan 14;122(2):e2418541122. doi: 10.1073/pnas.2418541122. Epub 2025 Jan 7. Proc Natl Acad Sci U S A. 2025. PMID: 39772737 - Regulation of Ribosome Biogenesis and Protein Synthesis Controls Germline Stem Cell Differentiation.
Sanchez CG, Teixeira FK, Czech B, Preall JB, Zamparini AL, Seifert JR, Malone CD, Hannon GJ, Lehmann R. Sanchez CG, et al. Cell Stem Cell. 2016 Feb 4;18(2):276-90. doi: 10.1016/j.stem.2015.11.004. Epub 2015 Dec 6. Cell Stem Cell. 2016. PMID: 26669894 Free PMC article. - Dimerisation of the PICTS complex via LC8/Cut-up drives co-transcriptional transposon silencing in Drosophila.
Eastwood EL, Jara KA, Bornelöv S, Munafò M, Frantzis V, Kneuss E, Barbar EJ, Czech B, Hannon GJ. Eastwood EL, et al. Elife. 2021 Feb 4;10:e65557. doi: 10.7554/eLife.65557. Elife. 2021. PMID: 33538693 Free PMC article.
References
- Brennecke J., Aravin A.A., Stark A., Dus M., Kellis M., Sachidanandam R., Hannon G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
- Chintapalli V.R., Wang J., Dow J.A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. - PubMed
- Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases