A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation - PubMed (original) (raw)
A ubiquitin-like domain recruits an oligomeric chaperone to a retrotranslocation complex in endoplasmic reticulum-associated degradation
Yue Xu et al. J Biol Chem. 2013.
Abstract
The Bag6-Ubl4A-Trc35 complex is a multifunctional chaperone that regulates various cellular processes. The diverse functions of Bag6 are supported by its ubiquitous localization to the cytoplasm, the nucleus, and membranes of the endoplasmic reticulum (ER) in cells. In ER-associated degradation (ERAD) pathways, Bag6 can interact with the membrane-associated ubiquitin ligase gp78 via its ubiquitin-like (UBL) domain, but the relative low affinity of this interaction does not reconcile with the fact that a fraction of Bag6 is tightly bound to the membranes. Here, we demonstrate that the UBL domain of Bag6 is required for interaction with the ER membranes. We find that in addition to gp78, the Bag6 UBL domain also binds a UBL-binding motif in UbxD8, an essential component of the gp78 ubiquitinating machinery. Importantly, Bag6 contains a proline-rich (PR) domain termed PDP (Proline rich-DUF3587-Proline rich) that forms homo-oligomer, allowing the UBL domain to form multivalent interactions with gp78 and UbxD8, which are essential for recruitment of Bag6 to the ER membrane. Furthermore, the PR domain comprises largely intrinsically disordered segments, which are sufficient for interaction with an unfolded substrate. We propose that simultaneous association with multiple ERAD factors helps to anchor a disordered chaperone oligomer to the site of retrotranslocation to prevent protein aggregation in ERAD.
Keywords: Bag6/Bat3/Scythe; E3 Ubiquitin Ligase; ER-associated Degradation; Endoplasmic Reticulum Stress; Endoplasmic Reticulum(ER); Holdase; Molecular Chaperone; Retrotranslocation; gp78.
Figures
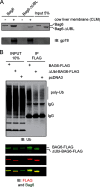
FIGURE 1.
The Bag6 UBL domain is required for membrane interaction. A, purified Bag6 or the Bag6ΔUBL mutant was incubated with salt-treated cow liver microsome membranes (CLM). The samples were layered on top of 1
m
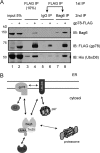
sucrose cushion and subject to centrifugation at 100,000 × g for 20 min. A fraction of the purified proteins and the proteins bound to the membrane were analyzed by immunoblotting (IB). Note that a small amount of Bag6 was present in the pellet fraction in the absent of CLM, likely due to oligomerization as described below. B, UBL domain is required for association with ubiquitinated proteins in cells. Cells transfected with the indicated DNA were lysed in a Nonidet P-40-containing buffer. A fraction of the extracts were analyzed directly by immunoblotting (input). The remaining samples were subject to immunoprecipitation with anti-FLAG antibodies. Bag6 and associated proteins were analyzed by immunoblotting.
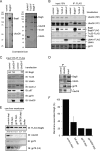
FIGURE 2.
Bag6 associates with the ER membrane via binding the UbxD8-gp78 complex. A, purification of Bag6, Trc35, or Ubl4A was performed using HEK293 cells expressing the indicated FLAG-tagged proteins. As a negative control, cells transfected with a control empty vector were used. The purified complexes were analyzed by SDS-PAGE and Coomassie Blue staining. B, immunoblotting confirms the interaction of Bag6 with UbxD8. Cells transfected with the indicated plasmids were lysed, and proteins immunoprecipitated with FLAG beads were analyzed by immunoblotting. LE, long exposure; S.E., short exposure. C, as in B, except that cells expressing FLAG-tagged UbxD8 were used. D, interaction of the endogenous Bag6 complex with UbxD8. Whole cell extracts were subject to immunoprecipitation by the indicated antibodies. The asterisk indicates IgG. E and F, UbxD8-gp78 complex is required for membrane association of Bag6. E, CLM extracts were treated with protein A beads containing either control IgG or the indicated antibodies. After depletion, proteolipisomes were re-formed and used in binding experiments with purified Bag6. Proteins bound to the membrane pellet fractions were analyzed by immunoblotting. Where indicated, a buffer control was included to assess the levels of background binding to residual Bio-Beads present in the samples. The numbers indicate band intensity. LE, long exposure. F, graph shows the quantification of the experiment in E. Error bar indicates the mean of the two independent experiments.
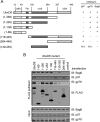
FIGURE 3.
The UBL domain of Bag6 is required for interaction with UbxD8. A, the domain structure of Bag6. B, cells were transfected with plasmids expressing the indicated Bag6 variants. The interactions of these Bag6 variants with Trc35, Ubl4A, and UbxD8 were analyzed by immunoprecipitation and immunoblotting.
FIGURE 4.
The UBA domain of UbxD8 interacts with Bag6. A, schematic representation of the UbxD8 constructs used in the interaction studies. The table summarizes the co-IP results. N.A., data not available due to a protein stability issue. B, cells expressing the indicated UbxD8 variants were lysed in the Nonidet P-40 lysis buffer. Immunoprecipitation was performed using FLAG beads.
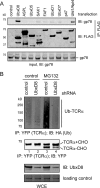
FIGURE 5.
UbxD8 is an essential co-factor of gp78. A, cells transfected with plasmids expressing the indicated UBX-containing proteins were lysed in the Nonidet P-40 lysis buffer. The UBX-containing proteins were immunopurified with FLAG beads. The purified proteins complexes were analyzed by immunoblotting with the indicated antibodies. B, cells stably expressing TCRα-YFP were transfected with either control or UbxD8-specific shRNA plasmid together with a construct expressing hemagglutinin (HA)-tagged ubiquitin. The cells were treated with either DMSO as control or with the proteasome inhibitor MG132 (10 μ
m
, 12 h). TCRα was immunoprecipitated from the cell extracts under a denaturing condition for analysis by immunoblotting. WCE, whole cell extract.
FIGURE 6.
Bag6 binds gp78 and UbxD8 simultaneously. A, cells expressing Bag6, UbxD8 in the presence or absence of gp78-FLAG were lysed in the Nonidet P-40 lysis buffer. The lysates were first subject to immunoprecipitation with FLAG beads to pull down gp78-FLAG. The bound materials eluted with FLAG peptide were divided into two equal portions. One potion was incubated with control IgG whereas the other portion was incubated with anti-Bag6 antibodies to purify Bag6 and its associated proteins. The precipitated materials and a fraction of the inputs were analyzed by immunoblotting. B, diagram illustrating interactions between Bag6 and various ERAD factors. Solid arrows indicate direct interactions whereas dashed lines indicate association that may be mediated by an adaptor.
FIGURE 7.
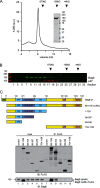
Bag6 forms a large oligomer. A, size exclusion chromatography analysis of purified Bag6. The Coomassie Blue-stained gel shows the purified protein. B, size exclusion chromatography analysis of whole cell extract. The fractions were analyzed by immunoblotting. C, a proline-rich (PR) domain is required for Bag6 oligomerization. The scheme shows the Bag6 constructs used in the interaction studies. Cells expressing the indicated Bag6 variants were lysed in Nonidet P-40 lysis buffer. Bag6 immunoprecipitated with the FLAG beads was analyzed by immunoblotting.
FIGURE 8.
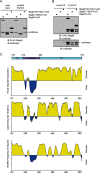
Bag6 employs a disordered domain to bind an unfolded protein. A, the PDP domain can prevent luciferase aggregation. Luciferase was incubated with excess amount of the indicated Bag6 variants at 42 °C for 20 min. A fraction of the samples were analyzed directly by immunoblotting (total input) whereas the remaining samples were subject to centrifugation to remove insoluble materials. A fraction of the soluble fractions were analyzed by immunoblotting. B, remaining samples of the soluble fractions were subject to immunoprecipitation with FLAG antibodies. The bound proteins were analyzed by immunoblotting. C, substrate binding domain of Bag6 is formed largely by disordered segments. The amino acid sequences of the Bag6 PDP domain were analyzed by the following secondary structure prediction programs, PONDR (top panel), IUPred (middle panel), and PONN (bottom panel).
Similar articles
- gp78 functions downstream of Hrd1 to promote degradation of misfolded proteins of the endoplasmic reticulum.
Zhang T, Xu Y, Liu Y, Ye Y. Zhang T, et al. Mol Biol Cell. 2015 Dec 1;26(24):4438-50. doi: 10.1091/mbc.E15-06-0354. Epub 2015 Sep 30. Mol Biol Cell. 2015. PMID: 26424800 Free PMC article. - USP13 antagonizes gp78 to maintain functionality of a chaperone in ER-associated degradation.
Liu Y, Soetandyo N, Lee JG, Liu L, Xu Y, Clemons WM Jr, Ye Y. Liu Y, et al. Elife. 2014;3:e01369. doi: 10.7554/eLife.01369. Epub 2014 Jan 14. Elife. 2014. PMID: 24424410 Free PMC article. - SGTA recognizes a noncanonical ubiquitin-like domain in the Bag6-Ubl4A-Trc35 complex to promote endoplasmic reticulum-associated degradation.
Xu Y, Cai M, Yang Y, Huang L, Ye Y. Xu Y, et al. Cell Rep. 2012 Dec 27;2(6):1633-44. doi: 10.1016/j.celrep.2012.11.010. Epub 2012 Dec 13. Cell Rep. 2012. PMID: 23246001 Free PMC article. - gp78: a multifaceted ubiquitin ligase that integrates a unique protein degradation pathway from the endoplasmic reticulum.
Chen Z, Du S, Fang S. Chen Z, et al. Curr Protein Pept Sci. 2012 Aug;13(5):414-24. doi: 10.2174/138920312802430590. Curr Protein Pept Sci. 2012. PMID: 22812524 Review. - The Gp78 ubiquitin ligase: probing endoplasmic reticulum complexity.
St Pierre P, Nabi IR. St Pierre P, et al. Protoplasma. 2012 Feb;249 Suppl 1:S11-8. doi: 10.1007/s00709-011-0344-8. Epub 2011 Nov 3. Protoplasma. 2012. PMID: 22045301 Review.
Cited by
- The effects of proteasomal inhibition on synaptic proteostasis.
Hakim V, Cohen LD, Zuchman R, Ziv T, Ziv NE. Hakim V, et al. EMBO J. 2016 Oct 17;35(20):2238-2262. doi: 10.15252/embj.201593594. Epub 2016 Sep 9. EMBO J. 2016. PMID: 27613546 Free PMC article. - The N-terminal Region of the Ubiquitin Regulatory X (UBX) Domain-containing Protein 1 (UBXD1) Modulates Interdomain Communication within the Valosin-containing Protein p97.
Trusch F, Matena A, Vuk M, Koerver L, Knævelsrud H, Freemont PS, Meyer H, Bayer P. Trusch F, et al. J Biol Chem. 2015 Dec 4;290(49):29414-27. doi: 10.1074/jbc.M115.680686. Epub 2015 Oct 16. J Biol Chem. 2015. PMID: 26475856 Free PMC article. - Digested disorder: Quarterly intrinsic disorder digest (April-May-June, 2013).
DeForte S, Reddy KD, Uversky VN. DeForte S, et al. Intrinsically Disord Proteins. 2013 Jan 1;1(1):e27454. doi: 10.4161/idp.27454. eCollection 2013 Jan-Dec. Intrinsically Disord Proteins. 2013. PMID: 28516028 Free PMC article. Review. - Structure of a BAG6 (Bcl-2-associated athanogene 6)-Ubl4a (ubiquitin-like protein 4a) complex reveals a novel binding interface that functions in tail-anchored protein biogenesis.
Kuwabara N, Minami R, Yokota N, Matsumoto H, Senda T, Kawahara H, Kato R. Kuwabara N, et al. J Biol Chem. 2015 Apr 10;290(15):9387-98. doi: 10.1074/jbc.M114.631804. Epub 2015 Feb 20. J Biol Chem. 2015. PMID: 25713138 Free PMC article. - BAG-6, a jack of all trades in health and disease.
Binici J, Koch J. Binici J, et al. Cell Mol Life Sci. 2014 May;71(10):1829-37. doi: 10.1007/s00018-013-1522-y. Epub 2013 Dec 4. Cell Mol Life Sci. 2014. PMID: 24305946 Free PMC article. Review.
References
- Ron D., Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8, 519–529 - PubMed
- Tsai B., Ye Y., Rapoport T. A. (2002) Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 3, 246–255 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials