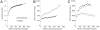Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage - PubMed (original) (raw)
Paul C Blainey, Senthil K Murugapiran, Wesley D Swingley, Christian A Ross, Susannah G Tringe, Patrick S G Chain, Matthew B Scholz, Chien-Chi Lo, Jason Raymond, Stephen R Quake, Brian P Hedlund
Affiliations
- PMID: 23673639
- PMCID: PMC3878185
- DOI: 10.1038/ncomms2884
Single-cell and metagenomic analyses indicate a fermentative and saccharolytic lifestyle for members of the OP9 lineage
Jeremy A Dodsworth et al. Nat Commun. 2013.
Abstract
OP9 is a yet-uncultivated bacterial lineage found in geothermal systems, petroleum reservoirs, anaerobic digesters and wastewater treatment facilities. Here we use single-cell and metagenome sequencing to obtain two distinct, nearly complete OP9 genomes, one constructed from single cells sorted from hot spring sediments and the other derived from binned metagenomic contigs from an in situ-enriched cellulolytic, thermophilic community. Phylogenomic analyses support the designation of OP9 as a candidate phylum for which we propose the name 'Atribacteria'. Although a plurality of predicted proteins is most similar to those from Firmicutes, the presence of key genes suggests a diderm cell envelope. Metabolic reconstruction from the core genome suggests an anaerobic lifestyle based on sugar fermentation by Embden-Meyerhof glycolysis with production of hydrogen, acetate and ethanol. Putative glycohydrolases and an endoglucanase may enable catabolism of (hemi)cellulose in thermal environments. This study lays a foundation for understanding the physiology and ecological role of the 'Atribacteria'.
Figures
Figure 1
Chimera-filtering increases contig N50 in the cSCG assembly. Stepwise assembly of composite OP9 SCG before (grey points) or after (black points) filtering out potentially chimeric reads. The increase in (A) total assembly size, (B) number of contigs, and (C) contig N50 are shown as a function of the number of reads (sampling with replacement) used.
Figure 2
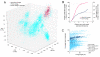
Identification of OP9-like contigs from the 77CS metagenome. (A) Principal component analysis (PCA) of tetranucleotide frequency of contigs in the 77CS metagenome and ten individual OP9 SCG assemblies. 77CS contigs in dark blue were binned as OP9-like, as defined by their placement within an ellipsoid with a centroid and semi-axis lengths equal to the mean and 2.5 times the standard deviation, respectively, of the first three principle components (PC1-PC3, with the percent variation explained by each in parentheses) of the OP9 SCG contigs. Only contigs greater than 2 kb in length are shown. Contigs <2kb inside this ellipsoid were also included in the OP9 bin if they had significant nucleotide identity (>85% identity over >100 nt) to contigs in the OP9-cSCG by BLASTn. (B) Total length of OP9 SCG and 77CS contigs >2kb contained within ellipsoids with different semi-axis lengths defined by multiplication of the standard deviation of PC1-PC3 of the OP9 SCG contigs by increasing scalar quantities (x-axis). Black points on the curves indicate the multiplier (2.5) used to define the OP9 bin, chosen at an approximate inflection point of the CS77 contig length curve to maximize inclusion of metagenome contigs in the cluster overlapping the OP9 SCGs but minimize inclusion of contigs in adjacent clusters. (C) Plot of coverage depth vs. contig length for the CS77 metagenome assembly, highlighting contigs in the OP9 bin with (dark blue circles) and without (open circles) >85% nucleotide identity over >100 bp to the OP9-cSCG assembly.
Figure 3
Relationship of OP9-SCG and OP9-CS77 to other bacterial groups. (A) Neighbor-joining tree based on a distance matrix of 1349 aligned positions of 16S rRNA genes from selected bacterial phyla with cultured representatives and the candidate phyla JS1 and OP9, including those in the OP9-SCG and OP9-CS77 assemblies. Black (100%), grey (>80%), and white (>50%) circles indicate bootstrap support (100 pseudoreplicates) for selected nodes in phylogenies inferred using neighbor-joining (left half of circle) and maximum-likelihood (right half) methods. (B) Maximum-likelihood phylogeny inferred from concatenated alignments of predicted amino acid sequences of 31 housekeeping genes identified by AMPHROA in genomes representing a variety of bacterial phyla and the OP9-SCG and OP9-CS77 assemblies. The number of genomes represented in each wedge is indicated in parentheses, and black circles indicate bootstrap support of >80% for 100 pseudoreplicates. (C) Binning of CDSs in the OP9 assemblies based on the phylogenetic affiliation of their top BLASTP hit to a database of sequenced bacterial and archaeal genomes. Phylogenetic groups with fewer than 1% of top hits were aggregated (‘other groups’). (D) Phylogenetic binning of CDSs on contigs or scaffolds containing markers diagnostic for a diderm cell envelope structure, emphasizing that top BLASTP hits were distributed among both monoderm and diderm Bacteria. Diderm Firmicutes includes members of the Negativicutes and Halanaerobiales.
Figure 4
Overview of features and potential metabolic capabilities of the OP9 lineage represented by the OP9-SCG and OP9-CS77 genomes as discussed in the text. CDSs in the OP9-cSCG and OP9-77CS associated with predicted functions are listed in Supplementary Table S5. Proteins involved in specific processes are identified by color: secretion and transporters (purple); saccharide catabolism and fermentation (red); energy conservation (orange); flagellar motility and chemotaxis (green); and nitrogen transport and assimilation (blue). Substrates and products are not necessarily balanced in the reactions depicted. Abbreviations not indicated in the text: ABC, ATP-binding cassette; Fdox, oxidized ferredoxin; AK, acetate kinase; PTA, phosphotransacetylase; AlDH, aldehyde dehydrogenase; ADH, alcohol dehydrogenase; PPi, pyrophosphate; Pi, inorganic phosphate.
Similar articles
- Phylogeny and physiology of candidate phylum 'Atribacteria' (OP9/JS1) inferred from cultivation-independent genomics.
Nobu MK, Dodsworth JA, Murugapiran SK, Rinke C, Gies EA, Webster G, Schwientek P, Kille P, Parkes RJ, Sass H, Jørgensen BB, Weightman AJ, Liu WT, Hallam SJ, Tsiamis G, Woyke T, Hedlund BP. Nobu MK, et al. ISME J. 2016 Feb;10(2):273-86. doi: 10.1038/ismej.2015.97. Epub 2015 Jun 19. ISME J. 2016. PMID: 26090992 Free PMC article. - Anaerobic hydrocarbon degradation in candidate phylum 'Atribacteria' (JS1) inferred from genomics.
Liu YF, Qi ZZ, Shou LB, Liu JF, Yang SZ, Gu JD, Mu BZ. Liu YF, et al. ISME J. 2019 Sep;13(9):2377-2390. doi: 10.1038/s41396-019-0448-2. Epub 2019 Jun 6. ISME J. 2019. PMID: 31171858 Free PMC article. - Genome-Resolved Metagenomic Analysis Reveals Roles for Candidate Phyla and Other Microbial Community Members in Biogeochemical Transformations in Oil Reservoirs.
Hu P, Tom L, Singh A, Thomas BC, Baker BJ, Piceno YM, Andersen GL, Banfield JF. Hu P, et al. mBio. 2016 Jan 19;7(1):e01669-15. doi: 10.1128/mBio.01669-15. mBio. 2016. PMID: 26787827 Free PMC article. - Genomic Characterization of Candidate Division LCP-89 Reveals an Atypical Cell Wall Structure, Microcompartment Production, and Dual Respiratory and Fermentative Capacities.
Youssef NH, Farag IF, Hahn CR, Jarett J, Becraft E, Eloe-Fadrosh E, Lightfoot J, Bourgeois A, Cole T, Ferrante S, Truelock M, Marsh W, Jamaleddine M, Ricketts S, Simpson R, McFadden A, Hoff W, Ravin NV, Sievert S, Stepanauskas R, Woyke T, Elshahed M. Youssef NH, et al. Appl Environ Microbiol. 2019 May 2;85(10):e00110-19. doi: 10.1128/AEM.00110-19. Print 2019 May 15. Appl Environ Microbiol. 2019. PMID: 30902854 Free PMC article. - Uncultivated thermophiles: current status and spotlight on 'Aigarchaeota'.
Hedlund BP, Murugapiran SK, Alba TW, Levy A, Dodsworth JA, Goertz GB, Ivanova N, Woyke T. Hedlund BP, et al. Curr Opin Microbiol. 2015 Jun;25:136-45. doi: 10.1016/j.mib.2015.06.008. Epub 2015 Jun 23. Curr Opin Microbiol. 2015. PMID: 26113243 Review.
Cited by
- Municipal Solid Waste Landfills Harbor Distinct Microbiomes.
Stamps BW, Lyles CN, Suflita JM, Masoner JR, Cozzarelli IM, Kolpin DW, Stevenson BS. Stamps BW, et al. Front Microbiol. 2016 Apr 20;7:534. doi: 10.3389/fmicb.2016.00534. eCollection 2016. Front Microbiol. 2016. PMID: 27148222 Free PMC article. - Single-Cell Genomics of Novel Actinobacteria With the Wood-Ljungdahl Pathway Discovered in a Serpentinizing System.
Merino N, Kawai M, Boyd ES, Colman DR, McGlynn SE, Nealson KH, Kurokawa K, Hongoh Y. Merino N, et al. Front Microbiol. 2020 Jun 9;11:1031. doi: 10.3389/fmicb.2020.01031. eCollection 2020. Front Microbiol. 2020. PMID: 32655506 Free PMC article. - Molecular Ecology of Hypersaline Microbial Mats: Current Insights and New Directions.
Wong HL, Ahmed-Cox A, Burns BP. Wong HL, et al. Microorganisms. 2016 Jan 5;4(1):6. doi: 10.3390/microorganisms4010006. Microorganisms. 2016. PMID: 27681900 Free PMC article. Review. - The majority of microorganisms in gas hydrate-bearing subseafloor sediments ferment macromolecules.
Zhang C, Fang YX, Yin X, Lai H, Kuang Z, Zhang T, Xu XP, Wegener G, Wang JH, Dong X. Zhang C, et al. Microbiome. 2023 Mar 2;11(1):37. doi: 10.1186/s40168-023-01482-5. Microbiome. 2023. PMID: 36864529 Free PMC article. - Single-Cell-Genomics-Facilitated Read Binning of Candidate Phylum EM19 Genomes from Geothermal Spring Metagenomes.
Becraft ED, Dodsworth JA, Murugapiran SK, Ohlsson JI, Briggs BR, Kanbar J, De Vlaminck I, Quake SR, Dong H, Hedlund BP, Swingley WD. Becraft ED, et al. Appl Environ Microbiol. 2015 Dec 4;82(4):992-1003. doi: 10.1128/AEM.03140-15. Print 2016 Feb 15. Appl Environ Microbiol. 2015. PMID: 26637598 Free PMC article.
References
- Rappé MS, Giovannoni SJ. The uncultured microbial majority. Ann. Rev. Microbiol. 2003;57:369–394. - PubMed
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 2008;6:431–440. - PubMed
- Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat. Rev. Microbiol. 2012;10:631–640. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources