Variation and genetic control of protein abundance in humans - PubMed (original) (raw)
Variation and genetic control of protein abundance in humans
Linfeng Wu et al. Nature. 2013.
Abstract
Gene expression differs among individuals and populations and is thought to be a major determinant of phenotypic variation. Although variation and genetic loci responsible for RNA expression levels have been analysed extensively in human populations, our knowledge is limited regarding the differences in human protein abundance and the genetic basis for this difference. Variation in messenger RNA expression is not a perfect surrogate for protein expression because the latter is influenced by an array of post-transcriptional regulatory mechanisms, and, empirically, the correlation between protein and mRNA levels is generally modest. Here we used isobaric tag-based quantitative mass spectrometry to determine relative protein levels of 5,953 genes in lymphoblastoid cell lines from 95 diverse individuals genotyped in the HapMap Project. We found that protein levels are heritable molecular phenotypes that exhibit considerable variation between individuals, populations and sexes. Levels of specific sets of proteins involved in the same biological process covary among individuals, indicating that these processes are tightly regulated at the protein level. We identified cis-pQTLs (protein quantitative trait loci), including variants not detected by previous transcriptome studies. This study demonstrates the feasibility of high-throughput human proteome quantification that, when integrated with DNA variation and transcriptome information, adds a new dimension to the characterization of gene expression regulation.
Figures
Fig. 1. Overview of workflow and protein association with ethnicity
a) Flow chart of experimental scheme. In each experiment, peptide digests from a reference cell line (GM12878) and five other cell lines were each labeled with one of the TMT-sixplex tags. Labeled peptides were equally mixed and subjected to identification and quantification by mass spectrometry, and then used for protein quantification. A total of 51 experiments were performed. b) The P value distribution for the difference in protein levels between CEU and YRI shows enrichment at small P values. c) P value of protein level differences between CEU and YRI plotted as a function of the genomic coordinate for each protein. The dashed line is at significance threshold Bonferroni P = 0.05. All the proteins that passed the threshold are highlighted with larger dots and labeled with gene names. Proteins that differed between CEU and YRI are distributed throughout the genome.
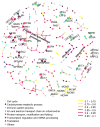
Fig. 2. Protein covariation network generated by sparse partial correlation estimation
Nodes represent proteins. Edges represent connection by covariation. This sparse network displays the 223 strongest connections among 278 proteins. Protein function was annotated by node color. Edge color was categorized according to correlation value. Known protein-protein interacting pairs were highlighted in larger nodes and labeled with gene names.
Fig. 3. Loci associated with protein expression levels
a) Identification of cis-pQTLs in all three populations combined (n=72). The P value and genomic coordinates for each protein/cis-SNP association test were plotted in the Manhattan plot. pQTLs with max(T) corrected P value < 0.001 were highlighted with a bigger dot size and a black outline. Multiple loci throughout the genome displayed an excess of small P values. Arrow indicates the location of the IMPA1 gene which contains a significant cis-pQTL. b) Overview of IMPA1 protein level and SNP genotype association in CEU, YRI, and all populations combined. The bottom plot is the fine mapping of cis-pQTL for IMPA1 based on HapMap I, II and III genotypes release 28. Each dot represents a tested SNP. Dot colors represent testing groups. The arrow is indicative of the chromosome location and transcription direction of the IMPA1 gene. There are several highly significant associations near the IMPA1 region in CEU and all populations combined. The exact locations of these associations in the IMPA1 gene region are illustrated in the top plot. The most significant SNP is rs1058401, located in IMPA1 3′UTR. c) Validation of IMPA1 protein expression level. IMPA1 protein expression level was validated by immunoblotting in 11 CEU individuals, with their genotype at rs1058401 labeled at the bottom. d) The bar plots show the mean of IMPA1 protein level of these 11 individuals in each rs1058401 genotype, based on data measured by quantitative mass spectrometry and by densitometry of immunoblot blots. Error bar, standard error of the mean. M.S., mass spectrometry. Im., immunoblotting.
Similar articles
- Defining the consequences of genetic variation on a proteome-wide scale.
Chick JM, Munger SC, Simecek P, Huttlin EL, Choi K, Gatti DM, Raghupathy N, Svenson KL, Churchill GA, Gygi SP. Chick JM, et al. Nature. 2016 Jun 23;534(7608):500-5. doi: 10.1038/nature18270. Epub 2016 Jun 15. Nature. 2016. PMID: 27309819 Free PMC article. - Identification and validation of genetic variants that influence transcription factor and cell signaling protein levels.
Hause RJ, Stark AL, Antao NN, Gorsic LK, Chung SH, Brown CD, Wong SS, Gill DF, Myers JL, To LA, White KP, Dolan ME, Jones RB. Hause RJ, et al. Am J Hum Genet. 2014 Aug 7;95(2):194-208. doi: 10.1016/j.ajhg.2014.07.005. Epub 2014 Jul 31. Am J Hum Genet. 2014. PMID: 25087611 Free PMC article. - Genetic control of the human brain proteome.
Robins C, Liu Y, Fan W, Duong DM, Meigs J, Harerimana NV, Gerasimov ES, Dammer EB, Cutler DJ, Beach TG, Reiman EM, De Jager PL, Bennett DA, Lah JJ, Wingo AP, Levey AI, Seyfried NT, Wingo TS. Robins C, et al. Am J Hum Genet. 2021 Mar 4;108(3):400-410. doi: 10.1016/j.ajhg.2021.01.012. Epub 2021 Feb 10. Am J Hum Genet. 2021. PMID: 33571421 Free PMC article. - Genetical genomics: combining genetics with gene expression analysis.
Li J, Burmeister M. Li J, et al. Hum Mol Genet. 2005 Oct 15;14 Spec No. 2:R163-9. doi: 10.1093/hmg/ddi267. Hum Mol Genet. 2005. PMID: 16244315 Review. - mRNAs, proteins and the emerging principles of gene expression control.
Buccitelli C, Selbach M. Buccitelli C, et al. Nat Rev Genet. 2020 Oct;21(10):630-644. doi: 10.1038/s41576-020-0258-4. Epub 2020 Jul 24. Nat Rev Genet. 2020. PMID: 32709985 Review.
Cited by
- Quantifying ubiquitin signaling.
Ordureau A, Münch C, Harper JW. Ordureau A, et al. Mol Cell. 2015 May 21;58(4):660-76. doi: 10.1016/j.molcel.2015.02.020. Mol Cell. 2015. PMID: 26000850 Free PMC article. Review. - Mapping of promoter usage QTL using RNA-seq data reveals their contributions to complex traits.
Kubota N, Suyama M. Kubota N, et al. PLoS Comput Biol. 2022 Aug 29;18(8):e1010436. doi: 10.1371/journal.pcbi.1010436. eCollection 2022 Aug. PLoS Comput Biol. 2022. PMID: 36037215 Free PMC article. - Longitudinal multi-omics study reveals common etiology underlying association between plasma proteome and BMI trajectories in adolescent and young adult twins.
Drouard G, Hagenbeek FA, Whipp AM, Pool R, Hottenga JJ, Jansen R, Hubers N, Afonin A; BIOS Consortium, BBMRI-N. L. Metabolomics Consortium; Willemsen G, de Geus EJC, Ripatti S, Pirinen M, Kanninen KM, Boomsma DI, van Dongen J, Kaprio J. Drouard G, et al. BMC Med. 2023 Dec 21;21(1):508. doi: 10.1186/s12916-023-03198-7. BMC Med. 2023. PMID: 38129841 Free PMC article. - Sex-dependent and sex-independent regulatory systems of size variation in natural populations.
Okada H, Yagi R, Gardeux V, Deplancke B, Hafen E. Okada H, et al. Mol Syst Biol. 2019 Nov;15(11):e9012. doi: 10.15252/msb.20199012. Mol Syst Biol. 2019. PMID: 31777173 Free PMC article. - Disentangling Genetic and Environmental Effects on the Proteotypes of Individuals.
Romanov N, Kuhn M, Aebersold R, Ori A, Beck M, Bork P. Romanov N, et al. Cell. 2019 May 16;177(5):1308-1318.e10. doi: 10.1016/j.cell.2019.03.015. Epub 2019 Apr 25. Cell. 2019. PMID: 31031010 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials