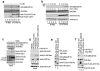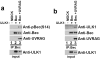ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase - PubMed (original) (raw)
ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase
Ryan C Russell et al. Nat Cell Biol. 2013 Jul.
Abstract
Autophagy is the primary cellular catabolic program activated in response to nutrient starvation. Initiation of autophagy, particularly by amino-acid withdrawal, requires the ULK kinases. Despite its pivotal role in autophagy initiation, little is known about the mechanisms by which ULK promotes autophagy. Here we describe a molecular mechanism linking ULK to the pro-autophagic lipid kinase VPS34. Following amino-acid starvation or mTOR inhibition, the activated ULK1 phosphorylates Beclin-1 on Ser 14, thereby enhancing the activity of the ATG14L-containing VPS34 complexes. The Beclin-1 Ser 14 phosphorylation by ULK is required for full autophagic induction in mammals and this requirement is conserved in Caenorhabditis elegans. Our study reveals a molecular link from ULK1 to activation of the autophagy-specific VPS34 complex and autophagy induction.
Conflict of interest statement
Competing Financial Interest
The author(s) declare no competing financial interests.
Figures
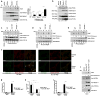
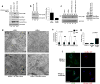
Fig.1. ULK is essential for activation of the ATG14L-associated VPS34 upon amino acid starvation
Unless otherwise stated experiments were repeated three times, data shown is representative. (a) Different VPS34 complexes were immunoprecipitated (IP) from MEF in the presence (N) or absence (-A) of amino acids using indicated antibodies and assayed for kinase activity (left, top panel). Inputs were immunoblotted using antibodies indicated (left, lower panels). Quantification of VPS34 activity is from 3 biological repeats (right panel, error bars denote S.D). (b) IP of three VPS34 complexes, normalized for VPS34, was performed using the indicated antibodies under nutrient rich or starvation conditions. VPS34 binding partners were analyzed by Western blot. (c) ATG14L-containing VPS34 complexes were immunopurified from wild-type, ULK1-/- 2KD (ULK def), and FIP200 -/- MEFs, and measured for lipid kinase activity similar to panel a. (d) VPS34 was immunopurified from wild-type, ULK def, and FIP200 -/- MEFs. Kinase assay was performed as in panel a. (e) IP of Beclin-1-containing VPS34 complexes from wild-type, ULK-def, and FIP200 -/- MEFs. Kinase assay was performed as in panel a. (f) LC3B puncta and PI3P levels were analyzed with anti-LC3B and Biotin-2XFYVE domain probe. Representative immunofluorescent images of LC3B and 2XFYVE domain binding were shown (scale bars,10μm). (g) Quantification of LC3B puncta from panel f. Details of quantification are provided in the methods section, data shown is mean -/+ S.D. Error bars in g-i are the standard deviation from a minimum of 6 unique fields of view from a representative experiment (see statistical source data). (h) Total PI3P was quantified from the experiment in panel f. Error bars was calculated as in g. (i) Quantification of PI3P that colocalizes with LC3B upon amino acid withdrawal from the experiment described in panel f. (j) VPS34 activity with or without ULK1 overexpression was assayed. A representative experiment of four repeats is shown. * in panels a,g,h,i denote a p-value <0.05 as determined by Student’s T-Test (see statistical source data and methods section). Quantification of PI3P/VPS34 provided under the PI3P panel in c-e,j nutrient rich conditions were normalized to 1.
Fig.2. Beclin-1 S14 is phosphorylated by ULK1 and required for VPS34 activation in response to amino acid withdrawal
Unless otherwise stated all experiments were repeated three times and data shown is representative. (a) HEK293 cells were transfected with ATG14L, VPS34, and Beclin-1. ATG14L-containing VPS34 complexes were immunopurified and subjected to an in vitro ULK1 kinase assay in the presence of 32-P-ATP. Bound ATG14L complexes and soluble ULK1 were separated and phosphorylation was detected by autoradiography (AR, left panels). Western blot was performed (right panels). Results are representative of two unique experiments. (b) Full length murine GST-Beclin-1 and various truncations (as labeled) were subjected to an in vitro HA-ULK1 kinase assay. GST-Beclin-1 6(S-T) A has serine-threonine residues 4,7,10,14,29,42 mutated to alanine. ULK1 inputs were determined by Western, Beclin-1 inputs by Coomassie (Coom) and target phosphorylation by AR. (c) GST-Beclin-1 (1-85) was subjected to an in vitro ULK1 kinase reaction and analyzed by mass spectrometry. S14, S15in humans, (boxed) is the major phosphorylation site identified (for mass spectrometry data, see Fig.S2b,c). Mass spectrometry was performed on a single experiment. (d) Beclin-1 S14 is the major in vitro ULK1phosphorylation site. Beclin-1 WT, S4A, and S14A mutant were subjected to an in vitro ULK1 kinase assay. The reaction developed by autoradiography and stained for Beclin-1 input levels by Coomassie stain. Results are representative of two unique experiments. (e) 293 cells were transfected with the indicated plasmids under nutrient rich conditions. Beclin-1 was IP’d and immunoblotted with pBeclin-1 (S14), or anti-Beclin-1 as a loading control. ULK1 inputs are included below IP samples. (f) Purified GST-Beclin-1 (1-85) was subjected to in vitro phosphorylation by GST-ULK1 (left panel) and GST-ULK2 (right panel). Reactions were immunoblotted with the indicated antibodies. (g) 293 cells were transfected with ATG14L, VPS34, and Beclin-1 and grown under nutrient rich conditions. ATG14L-containing VPS34 complexes were IP’d and lipid kinase activity was assayed as described in Fig.1j. Inputs were immunoblotted with the indicated antibodies. Representative of four unique experiments. (h) Stable lines containing Beclin1 (WT or S14A) were used for Beclin-1 IP. Binding partners were determined by SDS-PAGE analysis and Western blot using the indicated antibodies.
Fig.3. Beclin-1 is a physiological target of ULK kinase in response to amino acid withdrawal and mTOR inhibition
Unless otherwise stated all experiments were repeated three times and data shown is representative. (a) Wild-type MEF were cultured with or without amino acids. ATG14L-associated Beclin-1 was immunoprecipitated and treated with lambda phosphatase treatment (PPase) as indicated. Western blot was performed with the indicated antibodies. Beclin-1 S14 phosphorylation was quantified (shown under top panel) normalized to total Beclin-1. (b) Wild-type MEF were starved for the indicated time points. Beclin-1 was immunopurified by ATG14L IP and immunoblotted as indicated (top panels). Whole cell lysates were immunoblotted with pULK1 S757 (mTORC1-target site), pS6K and ULK1 antibodies (bottom panels). Two unique experiments were performed. (c) Wild-type or FIP200 -/- MEF were incubated under nutrient rich, amino acid deprived or Torin-1 (+T, an mTOR inhibitor) conditions. Beclin-1 was purified and immunoblotted as in Fig.3b. (d) Wild-type or ULK def MEF were incubated with or without amino acids. Beclin-1 was purified and immunoblotted as in Fig.3a. Two unique experiments were performed.
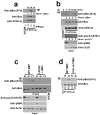
Fig.4. ATG14L stimulates Beclin-1 S14 phosphorylation by promoting association with ULK1
Unless otherwise stated all experiments were repeated three times and data shown is representative. (a) Beclin-1 alone (lanes 1-4) or Beclin-1 and ATG14L (lanes 5-8) were overexpressed in 293 cells. Beclin-1 was purified either by direct immunoprecipitation (lanes 1-4) or by ATG14L IP (lanes 5-8). IP samples were subjected to an in vitro ULK1 kinase assay with increasing amounts of ULK1. Reactions were immunoblotted with the indicated antibodies. Black line denotes discontinuous lanes from the same gel. Two unique experiments were performed. (b) Beclin-1 alone or bound to ATG14L was purified as described in panel a. Equal amounts of ULK1 were added to each complex and reactions were quenched at the indicated time points. Western blot was performed with the indicated antibodies. (c) An ATG14L-FLAG-6His inducible U2OS cell line was induced for 16 hours in the presence of amino acids. Endogenous Beclin-1 was immunoprecipitated and immunoblotted as in Fig.3a. ATG14L input levels are detected by immunoblotting. Two unique experiments were performed. (d) 293 cells transfected with either ATG14L or Beclin-1, or both, in conjunction with ULK1 were immunoprecipitated as indicated and blotted with the indicated antibodies. (e) 293 cells were transfected with Beclin-1 and ULK1 in the presence of ATG14L-WT or ATG14LΔCCD, which is defective in Beclin-1 binding, under nutrient rich conditions. Lysates were resolved by SDS-PAGE and blotted with the indicated antibodies. (f) 293 cells were transfected with ULK1 and Beclin-1 in conjunction with either ATG14L-WT, or one of two mutants (ΔBATS, ΔN) that are defective in phagophore localization. Samples were handled as in panel e.
Fig.5. UVRAG promotes Beclin-1 S14 phosphorylation and association with ULK1
(a) 293 cells were transfected with Beclin-1, with or without UVRAG, in conjunction with ULK1 as indicated in the presence of amino acids. Lysates were immunoblotted with the indicated antibodies. A representative experiment of three repeats is shown. (b) UVRAG bridges the interaction between Beclin-1 and ULK1. 293 cells were transfected with Beclin-1, with or without UVRAG, in conjunction with ULK1 as indicated. Lysates were immunoprecipitated with anti- HA(Beclin-1) antibody and blotted with the indicated antibodies. A representative experiment of three repeats is shown.
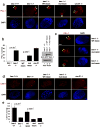
Fig.6. Beclin-1 S14 phosphorylation plays a critical role in autophagy induction by amino acid starvation
Unless otherwise stated all experiments were repeated three times and data shown is representative. (a) 293 cells were transfected with the indicated plasmids. Cells grown in the presence of amino acids and treated with NH4Cl to block autophagic turnover where indicated. Lysates were immunoblotted with the indicated antibodies. (b) 293 cells transfected with Beclin-1 ATG14L were grown in the presence or absence of amino acids. Lysates were immunoblotted with the indicated antibodies (left panel) and quantified by densitometry (right panel). Error bars represent the SD of three unique experiments. (c) shBeclin-1 reconstituted lines (wild-type or mutant) and controls (shScramble or shBeclin-1) were grown with or without amino acids were assessed for autophagy (left panel) and Beclin-1 levels (right panel). (d) Autophagosome (denoted by arrow heads) generation upon amino acid withdrawal was analyzed by electron microscopy. Cell lines and conditions from panel c were used and representative images from the indicated amino acid starved lines are shown; scale bar in bottom right represents 0.4μm. (e) Quantification of panel d. Fold induction was determined by arbitrarily making the nutrient rich condition 1 (solid bars) for each line. Error bars represent the standard deviation on the mean value over an average of 20 fields of view within a representative experiment. (f) HA-Beclin-1 WT or S14D was transiently expressed in FIP200 -/- MEF grown under nutrient rich conditions. Indirect immunofluorescence was performed using antibodies against endogenous LC3B and HA-Beclin-1. Scale bars represent 20μm. (g) Quantification of LC3B puncta from confocal displayed in panel f. In the HA-Beclin-1 or HA-Beclin-1-S14D transfected samples, only the HA staining positive cells were counted for LC3B puncta. Error bars were processed as in e. Mean value displayed, p-value determined by Student’s T-Test using 10 unique fields of view from panel f.
Fig.7. The conserved ULK phosphorylation site in C. elegans Bec-1 is required for autophagy
Unless otherwise stated all experiments were repeated three times and data shown is representative. (a) Bec-1 C. elegans were reconstituted with either wild-type or mutant GFP-BEC-1. Stable worm lines with Bec-1 rescue were obtained and embryos were stained with anti-PGL-1 antibody. Arrow indicates normal PGL-1 staining in germline cells. Scale bars represent 10μm. (b) Quantification of PGL-1 puncta outside germline cells (left panel). Error bars represents standard deviation between 3 unique embryos in a representative experiment. Reconstituted Bec-1 (WT and mut) levels in Bec -/- stable worms were compared by Western blot. Mean value presented. (c) Spectrum of defects in PGL granule degradation in bec-1 mutant rescue embryos. Mutant embryos displayed either high levels of diffuse PGL-1 staining (middle-left panel, 1/3 of the embryos), or large punctuate PGL-1 structures in somatic cells (bottom-left panel. 2/3 of the embryos). Both diffuse or punctuate PGL-1 staining in somatic cells have been described in autophagy deficient embryos. (d) Embryos from the lines described in Fig.6a were labeled with anti-LGG-1, along with wild-type and _unc_-51 worms. Representative embryos at ~100 cell stage are shown. (e) Quantification of LGG-1 per embryo from labeling in panel d. Error bars generated as in panel b. Mean value presented.
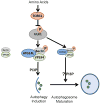
Fig.8. A working model of VPS34 complex regulation by ULK upon amino acid withdrawal
Amino acid starvation inactivates TORC1, de-repressing ULK1. ULK1 is recruited to VPS34-Beclin-1 complexes via binding to ATG14L, phosphorylating Beclin-1, activating the VPS34 kinase and PI3P production at the nascent autophagosome. Additionally, UVRAG-bound Beclin-1 is phosphorylated by ULK1, which may promote autophagosome maturation.
Comment in
- Autophagy: Kinase crosstalk through beclin 1.
Wrighton KH. Wrighton KH. Nat Rev Mol Cell Biol. 2013 Jul;14(7):402-3. doi: 10.1038/nrm3608. Epub 2013 Jun 12. Nat Rev Mol Cell Biol. 2013. PMID: 23756621 No abstract available. - ULK1 targets Beclin-1 in autophagy.
Nazarko VY, Zhong Q. Nazarko VY, et al. Nat Cell Biol. 2013 Jul;15(7):727-8. doi: 10.1038/ncb2797. Nat Cell Biol. 2013. PMID: 23817237 Free PMC article.
Similar articles
- ULK1 targets Beclin-1 in autophagy.
Nazarko VY, Zhong Q. Nazarko VY, et al. Nat Cell Biol. 2013 Jul;15(7):727-8. doi: 10.1038/ncb2797. Nat Cell Biol. 2013. PMID: 23817237 Free PMC article. - Cul3-KLHL20 Ubiquitin Ligase Governs the Turnover of ULK1 and VPS34 Complexes to Control Autophagy Termination.
Liu CC, Lin YC, Chen YH, Chen CM, Pang LY, Chen HA, Wu PR, Lin MY, Jiang ST, Tsai TF, Chen RH. Liu CC, et al. Mol Cell. 2016 Jan 7;61(1):84-97. doi: 10.1016/j.molcel.2015.11.001. Epub 2015 Dec 10. Mol Cell. 2016. PMID: 26687681 - The ULK1 complex mediates MTORC1 signaling to the autophagy initiation machinery via binding and phosphorylating ATG14.
Park JM, Jung CH, Seo M, Otto NM, Grunwald D, Kim KH, Moriarity B, Kim YM, Starker C, Nho RS, Voytas D, Kim DH. Park JM, et al. Autophagy. 2016;12(3):547-64. doi: 10.1080/15548627.2016.1140293. Autophagy. 2016. PMID: 27046250 Free PMC article. - So Many Roads: the Multifaceted Regulation of Autophagy Induction.
Corona Velazquez AF, Jackson WT. Corona Velazquez AF, et al. Mol Cell Biol. 2018 Oct 15;38(21):e00303-18. doi: 10.1128/MCB.00303-18. Print 2018 Nov 1. Mol Cell Biol. 2018. PMID: 30126896 Free PMC article. Review. - Autophagy regulation by nutrient signaling.
Russell RC, Yuan HX, Guan KL. Russell RC, et al. Cell Res. 2014 Jan;24(1):42-57. doi: 10.1038/cr.2013.166. Epub 2013 Dec 17. Cell Res. 2014. PMID: 24343578 Free PMC article. Review.
Cited by
- Therapeutic targeting of autophagy: potential and concerns in treating cardiovascular disease.
Orogo AM, Gustafsson ÅB. Orogo AM, et al. Circ Res. 2015 Jan 30;116(3):489-503. doi: 10.1161/CIRCRESAHA.116.303791. Circ Res. 2015. PMID: 25634972 Free PMC article. Review. - Autophagy protein ULK1 interacts with and regulates SARM1 during axonal injury.
Choi HMC, Li Y, Suraj D, Hsia RC, Sarkar C, Wu J, Lipinski MM. Choi HMC, et al. Proc Natl Acad Sci U S A. 2022 Nov 22;119(47):e2203824119. doi: 10.1073/pnas.2203824119. Epub 2022 Nov 14. Proc Natl Acad Sci U S A. 2022. PMID: 36375051 Free PMC article. - Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase.
Lin M, Liu H, Xiong Q, Niu H, Cheng Z, Yamamoto A, Rikihisa Y. Lin M, et al. Autophagy. 2016 Nov;12(11):2145-2166. doi: 10.1080/15548627.2016.1217369. Epub 2016 Aug 19. Autophagy. 2016. PMID: 27541856 Free PMC article. - USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1.
Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, Li X, Liang W, Lu X, Qian L, Yuan J. Xu D, et al. Genes Dev. 2016 Aug 1;30(15):1718-30. doi: 10.1101/gad.285122.116. Genes Dev. 2016. PMID: 27542828 Free PMC article. - Continuous administration of the mTORC1 inhibitor everolimus induces tolerance and decreases autophagy in mice.
Kurdi A, De Doncker M, Leloup A, Neels H, Timmermans JP, Lemmens K, Apers S, De Meyer GRY, Martinet W. Kurdi A, et al. Br J Pharmacol. 2016 Dec;173(23):3359-3371. doi: 10.1111/bph.13626. Epub 2016 Oct 23. Br J Pharmacol. 2016. PMID: 27638766 Free PMC article.
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. - PubMed
- Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. Journal of cell science. 2006;119:3888–3900. - PubMed
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- CA108941/CA/NCI NIH HHS/United States
- R01 CA132809/CA/NCI NIH HHS/United States
- CAPMC/ CIHR/Canada
- GM51586/GM/NIGMS NIH HHS/United States
- R01 GM062694/GM/NIGMS NIH HHS/United States
- R01 GM051586/GM/NIGMS NIH HHS/United States
- R01 CA108941/CA/NCI NIH HHS/United States
- GM62694/GM/NIGMS NIH HHS/United States
- R01 ES021667/ES/NIEHS NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous