Regulation of T cell function by the ubiquitin-specific protease USP9X via modulating the Carma1-Bcl10-Malt1 complex - PubMed (original) (raw)
Regulation of T cell function by the ubiquitin-specific protease USP9X via modulating the Carma1-Bcl10-Malt1 complex
Yoon Park et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The ubiquitin conjugation system plays an important role in immune regulation; however, the ubiquitin-specific proteases (USPs) that carry out deubiquitination of cellular substrates are poorly understood. Here we show that in vivo knockdown of the deubiquitinating enzyme USP9X attenuates T-cell proliferation. In addition, naïve CD4(+) T cells from USP9X knockdown chimeric mice display decreased cytokine production and T helper cell differentiation in vitro, which we confirmed in vivo by performing adoptive transfer of transgenic T cells and subsequent immunization. USP9X silencing in both a human T-cell line and mouse primary T cells reduced T-cell receptor (TCR) signaling-induced NF-κB activation. Mechanistically, USP9X interacts with Bcl10 of the Carma1-Bcl10-Malt1 (CBM) complex and removes the TCR-induced ubiquitin chain from Bcl10, which facilitates the association of Carma1 with Bcl0-Malt1. These results demonstrate that USP9X is a crucial positive regulator of the TCR signaling pathway and is required for T-cell function through the modulation of CBM complex formation.
Keywords: posttranslational modification; signal transduction.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Characterization of USP9X-deficient T cells from bone marrow chimeric mice. (A) Endogenous USP9X protein level was measured by immunoblotting with anti-USP9X antibody in various mouse tissues. The membrane was reprobed with anti-actin to reveal equivalent protein loading. (B) (Upper) Analysis of GFP expression in peripheral blood from bone marrow chimeric mice at 2 mo after reconstitution of USP9X shRNAs expressing bone marrow cells. (Lower) Immunoblot analysis of USP9X was performed in a sorted GFP+ CD4+ T-cell population. The data are representative of between 3 and 10 independent experiments. (C) CD4+ T cells were sorted from USP9X shRNAs expressing chimeric mice and stimulated with anti-CD3 + anti-CD28 for 36 h. Cell proliferation was measured by 3H-thymidine uptake. The data are compiled from between 3 and 10 independent experiments. Error bars indicate mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001, two-tailed unpaired t test. (D) Violet dye-labeled CD4+ T cells were stimulated with anti-CD3 + anti-CD28 for 2 and 4 d, after which cell division was analyzed by flow cytometry. The data are representative of three independent experiments. (E) Cell death of stimulated CD4+ T cells was examined by 7-AAD staining and analyzed by flow cytometry (Upper) and total cell counts (Lower). The data are representative of three independent experiments. (F) IL-2 production by OVA-specific CD4 T cells. CD4+ T cells from OT-II control or OT-II USP9X knockdown chimeric mice were adoptively transferred into WT C57BL/6J mice, followed by immunization with OVA plus CFA as an adjuvant. Splenocytes and lymph node cells obtained 6 d later were stimulated with OVA323–339 peptide for 24 h and analyzed by intracellular staining and flow cytometry (Upper) and ELISA (Lower). The data are representative of and compiled from two independent experiments (n = 2 per each experiment). Error bars indicate mean ± SD. *P < 0.05, two-tailed unpaired t test.
Fig. 2.
Defective Th cell differentiation of USP9X-deficient T cells. (A) Sorted GFP+CD4+CD62L+CD25− naïve CD4+ T cells from control (shCON) and USP9X shRNA (shUSP9X)-expressing chimeric mice were stimulated with anti-CD3 + anti-CD28 and cultured under different polarizing conditions for Th-neutral, Th1, Th2, or Th17 for 5 d. Cells producing IFN-γ, IL-4, and IL-17 were analyzed by intracellular cytokine staining (ICCS) at 6 h after restimulation with anti-CD3 + anti-CD28. The data are representative of five independent experiments. (B) IFN-γ, IL-4, and IL-17 production were measured by ELISA at 24 h after restimulation. The data are compiled from five independent experiments. Error bars indicate mean ± SD. *P < 0.05;**P < 0.01, two-tailed unpaired t test. (C and D) CD4+ T cells from OT-II control or OT-II USP9X knockdown chimeric mice were adoptively transferred into WT C57BL/6J mice, followed by immunization with OVA plus CFA (C) or alum (D) as adjuvants. Splenocytes and lymph node cells obtained 6 d later were stimulated and analyzed as described in Fig. 1_F_. The data are representative of three independent experiments. (E and F) Cytokine expression profile in C57BL/6J mice immunized as in C and D. CD4+ T cells obtained at 6 d after immunization were stimulated with OVA323–339 peptide for 48 h, and the culture supernatants were analyzed by a bioplex multicytokine assay. The data are compiled from three independent experiments. Error bars indicate mean ± SD. *P < 0.05; **P < 0.01, two-tailed unpaired t test.
Fig. 3.
Impaired TCR/CD28-induced NF-κB activation in USP9X-deficient T cells. (A) Analysis of the phosphorylation status of TCR signaling proteins. Control and USP9X shRNA-expressing JE6.1 cells were stimulated with anti-CD3 + anti-CD28 (α-CD3/28) for the indicated times, after which the cell lysates were subjected to immunoblotting with the indicated antibodies. Anti-actin blot was used as a loading control. (B) Immunoblot analysis of translocation of p65 to the nucleus in control and USP9X shRNA-expressing JE6.1 cells treated as in A. After stimulation, cell lysates were separated into nuclear and cytosolic fractions. (C) Immunoblot analysis of translocation of p65 to the nucleus in mouse CD4+ T cells. Sorted Naïve CD4+ T cells from control and USP9X knockdown chimeric mice were stimulated with anti-CD3 + anti-CD28 for the indicated times, and nuclear fractionation was performed as in B. The protein content of IκBα was detected in the total cell lysates (Lower). The data are representative of three independent experiments. (D) Immunoblot analysis of phosphorylation of IκBα in USP9X shRNA-expressing JTAg cells reconstituted with USP9X. USP9X shRNA-expressing JTAg cells were transfected with empty vector or USP9X and CD28 and stimulated with anti-CD3 + anti-CD28 for the indicated times, after which the cell lysates were subjected to immunoblotting with the indicated antibodies. The data are representative of two independent experiments.
Fig. 4.
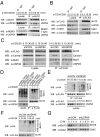
USP9X is required for CBM complex formation. (A) Interaction between endogenous USP9X and Bcl10. JE6.1 cells were stimulated with anti-CD3 + anti-CD28 for the indicated times, and endogenous Bcl10 or NEMO was immunoprecipitated with antibodies against Bcl10 or NEMO, respectively. The immunoprecipitates were separated by SDS/PAGE and immunoblotted with the indicated antibodies. (B) Interaction between USP9X and the CBM complex. JE6.1 cells expressing FLAG-Bcl10 were stimulated as in A, after which the FLAG-Bcl10 was immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were then subjected to SDS/PAGE and immunoblotted with the indicated antibodies. (C) JE6.1 cells stably expressing both FLAG-Bcl10 and control or USP9X shRNA were stimulated with anti-CD3 + anti-CD28 for the indicated times, after which FLAG-Bcl10 was immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were subjected to SDS/PAGE and immunoblotted with the indicated antibodies. (D) TCR-induced ubiquitination of Bcl10 in WT USP9X and catalytically inactive mutant (C1559A)-overexpressing cells. JTAg cells were transiently transfected with expression constructs for CD28, FLAG-Bcl10, HA ubiquitin, and USP9X WT or USP9X C1559A mutant. At 48 h after transfection, cells were left untreated or were stimulated with anti-CD3 + anti-CD28 for 20 min. Cell lysates were generated by adding SDS lysis buffer (2% SDS) and immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were subjected to SDS/PAGE and analyzed by immunoblotting with the indicated antibodies. (E) TCR-induced ubiquitination of Bcl10 in control and USP9X shRNA-expressing cells. JTAg cells stably expressing control or USP9X shRNA were transfected with FLAG-Bcl10, HA ubiquitin, and CD28 and stimulated with anti-CD3 + anti-CD28 for the indicated times, and then analyzed as in D. The data are representative of three independent experiments. (F) TCR-induced K48-linked ubiquitination of Bcl10 in control and USP9X-shRNA expressing cells. JTAg cells stably expressing control or USP9X shRNA were transfected with FLAG-Bcl10, HA-K48–only ubiquitin, and CD28; stimulated with anti-CD3 + anti-CD28 for the indicated times; and then analyzed as in D. The data are representative of three independent experiments. (G) Stability of Bcl10 in JE6.1 cells expressing USP9X shRNA. Cells were stimulated with anti-CD3 + anti-CD28 and cycloheximide (50 μg/mL) was added for the indicated times. The cell lysates were subjected to immunoblotting with the indicated antibodies. Anti-Grb2 blot was used as a loading control. The data are representative of two independent experiments.
Similar articles
- The Ca2+-dependent phosphatase calcineurin controls the formation of the Carma1-Bcl10-Malt1 complex during T cell receptor-induced NF-kappaB activation.
Palkowitsch L, Marienfeld U, Brunner C, Eitelhuber A, Krappmann D, Marienfeld RB. Palkowitsch L, et al. J Biol Chem. 2011 Mar 4;286(9):7522-34. doi: 10.1074/jbc.M110.155895. Epub 2011 Jan 3. J Biol Chem. 2011. PMID: 21199863 Free PMC article. - T cell receptor-dependent activation of mTOR signaling in T cells is mediated by Carma1 and MALT1, but not Bcl10.
Hamilton KS, Phong B, Corey C, Cheng J, Gorentla B, Zhong X, Shiva S, Kane LP. Hamilton KS, et al. Sci Signal. 2014 Jun 10;7(329):ra55. doi: 10.1126/scisignal.2005169. Sci Signal. 2014. PMID: 24917592 Free PMC article. - Role of the CARMA1/BCL10/MALT1 complex in lymphoid malignancies.
Juilland M, Thome M. Juilland M, et al. Curr Opin Hematol. 2016 Jul;23(4):402-9. doi: 10.1097/MOH.0000000000000257. Curr Opin Hematol. 2016. PMID: 27135977 Free PMC article. Review. - Protein kinase C-δ negatively regulates T cell receptor-induced NF-κB activation by inhibiting the assembly of CARMA1 signalosome.
Liu Y, Song R, Gao Y, Li Y, Wang S, Liu HY, Wang Y, Hu YH, Shu HB. Liu Y, et al. J Biol Chem. 2012 Jun 8;287(24):20081-7. doi: 10.1074/jbc.M111.335463. Epub 2012 Apr 23. J Biol Chem. 2012. PMID: 22528498 Free PMC article. - Critical protein-protein interactions within the CARMA1-BCL10-MALT1 complex: Take-home points for the cell biologist.
Cheng J, Maurer LM, Kang H, Lucas PC, McAllister-Lucas LM. Cheng J, et al. Cell Immunol. 2020 Sep;355:104158. doi: 10.1016/j.cellimm.2020.104158. Epub 2020 Jul 7. Cell Immunol. 2020. PMID: 32721634 Review.
Cited by
- USP28 protects development of inflammation in mouse intestine by regulating STAT5 phosphorylation and IL22 production in T lymphocytes.
Le Menn G, Pikkarainen K, Mennerich D, Miroszewska D, Kietzmann T, Chen Z. Le Menn G, et al. Front Immunol. 2024 Jul 15;15:1401949. doi: 10.3389/fimmu.2024.1401949. eCollection 2024. Front Immunol. 2024. PMID: 39076972 Free PMC article. - USP9X deubiquitylating enzyme maintains RAPTOR protein levels, mTORC1 signalling and proliferation in neural progenitors.
Bridges CR, Tan MC, Premarathne S, Nanayakkara D, Bellette B, Zencak D, Domingo D, Gecz J, Murtaza M, Jolly LA, Wood SA. Bridges CR, et al. Sci Rep. 2017 Mar 24;7(1):391. doi: 10.1038/s41598-017-00149-0. Sci Rep. 2017. PMID: 28341829 Free PMC article. - Emerging role of deubiquitination modifications of programmed death-ligand 1 in cancer immunotherapy.
Jiang Y, Hong K, Zhao Y, Xu K. Jiang Y, et al. Front Immunol. 2023 Jun 21;14:1228200. doi: 10.3389/fimmu.2023.1228200. eCollection 2023. Front Immunol. 2023. PMID: 37415977 Free PMC article. Review. - USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells.
Chen Z, Wang HW, Wang S, Fan L, Feng S, Cai X, Peng C, Wu X, Lu J, Chen D, Chen Y, Wu W, Lu D, Liu N, You Y, Wang H. Chen Z, et al. J Clin Invest. 2019 Apr 8;129(5):2043-2055. doi: 10.1172/JCI126414. eCollection 2019 Apr 8. J Clin Invest. 2019. PMID: 30958800 Free PMC article. - Post-translational Modifications of the CARMA1-BCL10-MALT1 Complex in Lymphocytes and Activated B-Cell Like Subtype of Diffuse Large B-Cell Lymphoma.
Thys A, Douanne T, Bidère N. Thys A, et al. Front Oncol. 2018 Nov 9;8:498. doi: 10.3389/fonc.2018.00498. eCollection 2018. Front Oncol. 2018. PMID: 30474008 Free PMC article. Review.
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. - PubMed
- Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. - PubMed
- Nijman SM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. - PubMed
- Komander D, Clague MJ, Urbé S. Breaking the chains: Structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–563. - PubMed
- Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430(7000):694–699. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI78272/AI/NIAID NIH HHS/United States
- S10 RR027366/RR/NCRR NIH HHS/United States
- R01 AI062969/AI/NIAID NIH HHS/United States
- R01 AI078272/AI/NIAID NIH HHS/United States
- R01 AI62969/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials