Topographic diversity of fungal and bacterial communities in human skin - PubMed (original) (raw)
. 2013 Jun 20;498(7454):367-70.
doi: 10.1038/nature12171. Epub 2013 May 22.
Collaborators, Affiliations
- PMID: 23698366
- PMCID: PMC3711185
- DOI: 10.1038/nature12171
Topographic diversity of fungal and bacterial communities in human skin
Keisha Findley et al. Nature. 2013.
Abstract
Traditional culture-based methods have incompletely defined the microbial landscape of common recalcitrant human fungal skin diseases, including athlete's foot and toenail infections. Skin protects humans from invasion by pathogenic microorganisms and provides a home for diverse commensal microbiota. Bacterial genomic sequence data have generated novel hypotheses about species and community structures underlying human disorders. However, microbial diversity is not limited to bacteria; microorganisms such as fungi also have major roles in microbial community stability, human health and disease. Genomic methodologies to identify fungal species and communities have been limited compared with those that are available for bacteria. Fungal evolution can be reconstructed with phylogenetic markers, including ribosomal RNA gene regions and other highly conserved genes. Here we sequenced and analysed fungal communities of 14 skin sites in 10 healthy adults. Eleven core-body and arm sites were dominated by fungi of the genus Malassezia, with only species-level classifications revealing fungal-community composition differences between sites. By contrast, three foot sites--plantar heel, toenail and toe web--showed high fungal diversity. Concurrent analysis of bacterial and fungal communities demonstrated that physiologic attributes and topography of skin differentially shape these two microbial communities. These results provide a framework for future investigation of the contribution of interactions between pathogenic and commensal fungal and bacterial communities to the maintainenace of human health and to disease pathogenesis.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
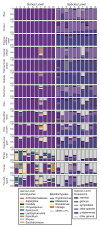
Figure 1. Relative abundance of fungal genera and Malassezia species of human skin sites
Fungal diversity of individual body sites of healthy volunteers (HV) was taxonomically classified at the genus level with further resolution of Malassezia species. Note: Left side is shown for all body sites except HV7’s right toenail.
Figure 2. Median richness of fungal and bacterial genera
Median taxonomic richness (or number of observed genera) of fungal and bacterial genera at 14 body sites. Error bars represent the median absolute deviation.
Figure 3. Forces shaping fungal and bacterial communities
Principal coordinates analysis (PCoA) of degree of fungal and bacterial community similarity at 14 body sites, based on predominant genera and species. Variation in fungal communities segregated strongly along site location, with feet, head, and torso sites forming discrete groups (A). Bacterial community structure was more dependent on site physiology (B). Axes that most significantly contribute to variation and their relative length are defined in Figure S10 legend. For fungi, M. restricta (i; ρ = 0.92) and M. globosa (a; ρ = −0.79) are primary and opposing drivers of variation. For bacteria, Propionibacterium (j; ρ = 0.95) contributes to sebaceous site variation, while Corynebacterium (l; ρ = −0.74), and Turicella (q; ρ = −0.56) contribute most heavily for moist sites.
Figure 4. Clinical involvement alters shared fungal community structure
Community structure measures the type and relative abundance of each genus. A value of 1 implies identical community structure and 0 implies dissimilar structures. Amongst uninvolved feet sites, community structure is fairly consistent at plantar heel, toenail and toeweb. For involved sites, plantar heel has much greater shared community structure and toenails have much lower shared community structure. Error bars represent the standard error of the mean.
Similar articles
- Dysbiotic Bacterial and Fungal Communities Not Restricted to Clinically Affected Skin Sites in Dandruff.
Soares RC, Camargo-Penna PH, de Moraes VC, De Vecchi R, Clavaud C, Breton L, Braz AS, Paulino LC. Soares RC, et al. Front Cell Infect Microbiol. 2016 Nov 17;6:157. doi: 10.3389/fcimb.2016.00157. eCollection 2016. Front Cell Infect Microbiol. 2016. PMID: 27909689 Free PMC article. - Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions.
Paulino LC, Tseng CH, Strober BE, Blaser MJ. Paulino LC, et al. J Clin Microbiol. 2006 Aug;44(8):2933-41. doi: 10.1128/JCM.00785-06. J Clin Microbiol. 2006. PMID: 16891514 Free PMC article. - Diverse Human Skin Fungal Communities in Children Converge in Adulthood.
Jo JH, Deming C, Kennedy EA, Conlan S, Polley EC, Ng WI; NISC Comparative Sequencing Program; Segre JA, Kong HH. Jo JH, et al. J Invest Dermatol. 2016 Dec;136(12):2356-2363. doi: 10.1016/j.jid.2016.05.130. Epub 2016 Jul 29. J Invest Dermatol. 2016. PMID: 27476723 Free PMC article. - Microbial Ecology of the Human Skin.
Cundell AM. Cundell AM. Microb Ecol. 2018 Jul;76(1):113-120. doi: 10.1007/s00248-016-0789-6. Epub 2016 May 31. Microb Ecol. 2018. PMID: 27245597 Review. - Topographical and physiological differences of the skin mycobiome in health and disease.
Jo JH, Kennedy EA, Kong HH. Jo JH, et al. Virulence. 2017 Apr 3;8(3):324-333. doi: 10.1080/21505594.2016.1249093. Epub 2016 Oct 18. Virulence. 2017. PMID: 27754756 Free PMC article. Review.
Cited by
- Estimating the quality of eukaryotic genomes recovered from metagenomic analysis with EukCC.
Saary P, Mitchell AL, Finn RD. Saary P, et al. Genome Biol. 2020 Sep 10;21(1):244. doi: 10.1186/s13059-020-02155-4. Genome Biol. 2020. PMID: 32912302 Free PMC article. - A genome catalog of the early-life human skin microbiome.
Shen Z, Robert L, Stolpman M, Che Y, Allen KJ, Saffery R, Walsh A, Young A, Eckert J, Deming C, Chen Q, Conlan S, Laky K, Li JM, Chatman L, Kashaf SS; NISC Comparative Sequencing Program; VITALITY team; Kong HH, Frischmeyer-Guerrerio PA, Perrett KP, Segre JA. Shen Z, et al. Genome Biol. 2023 Nov 10;24(1):252. doi: 10.1186/s13059-023-03090-w. Genome Biol. 2023. PMID: 37946302 Free PMC article. - Potential of Skin Microbiome, Pro- and/or Pre-Biotics to Affect Local Cutaneous Responses to UV Exposure.
Patra V, Gallais Sérézal I, Wolf P. Patra V, et al. Nutrients. 2020 Jun 17;12(6):1795. doi: 10.3390/nu12061795. Nutrients. 2020. PMID: 32560310 Free PMC article. Review. - Gut Fungal Microbiota Alterations in Pulmonary Arterial Hypertensive Rats.
Chen Y, Meng L, Yuan W, Gao Z, Zhang X, Xie B, Song J, Li J, Zhong J, Liu X. Chen Y, et al. Biomedicines. 2024 Jan 27;12(2):298. doi: 10.3390/biomedicines12020298. Biomedicines. 2024. PMID: 38397900 Free PMC article. - The cutaneous microbiome in outpatients presenting with acute skin abscesses.
Horton JM, Gao Z, Sullivan DM, Shopsin B, Perez-Perez GI, Blaser MJ. Horton JM, et al. J Infect Dis. 2015 Jun 15;211(12):1895-904. doi: 10.1093/infdis/jiv003. Epub 2015 Jan 12. J Infect Dis. 2015. PMID: 25583170 Free PMC article.
References
- Marples M, editor. The Ecology of the Human Skin. Bannerstone House; Springfield, Illinois: 1965.
- Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annu Rev Pathol. 2012;7:99–122. - PubMed
- Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. 2010;8:340–9. - PubMed
Publication types
MeSH terms
Grants and funding
- 1K99AR059222/AR/NIAMS NIH HHS/United States
- ZIA HG000180-12/Intramural NIH HHS/United States
- ZIA BC010938-05/Intramural NIH HHS/United States
- 4UH3AR057504-02/AR/NIAMS NIH HHS/United States
- 1UH2AR057504-01/AR/NIAMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases