BAF complexes facilitate decatenation of DNA by topoisomerase IIα - PubMed (original) (raw)
. 2013 May 30;497(7451):624-7.
doi: 10.1038/nature12146. Epub 2013 May 22.
Affiliations
- PMID: 23698369
- PMCID: PMC3668793
- DOI: 10.1038/nature12146
BAF complexes facilitate decatenation of DNA by topoisomerase IIα
Emily C Dykhuizen et al. Nature. 2013.
Abstract
Recent exon-sequencing studies of human tumours have revealed that subunits of BAF (mammalian SWI/SNF) complexes are mutated in more than 20% of all human malignancies, but the mechanisms involved in tumour suppression are unclear. BAF chromatin-remodelling complexes are polymorphic assemblies that use energy provided by ATP hydrolysis to regulate transcription through the control of chromatin structure and the placement of Polycomb repressive complex 2 (PRC2) across the genome. Several proteins dedicated to this multisubunit complex, including BRG1 (also known as SMARCA4) and BAF250a (also known as ARID1A), are mutated at frequencies similar to those of recognized tumour suppressors. In particular, the core ATPase BRG1 is mutated in 5-10% of childhood medulloblastomas and more than 15% of Burkitt's lymphomas. Here we show a previously unknown function of BAF complexes in decatenating newly replicated sister chromatids, a requirement for proper chromosome segregation during mitosis. We find that deletion of Brg1 in mouse cells, as well as the expression of BRG1 point mutants identified in human tumours, leads to anaphase bridge formation (in which sister chromatids are linked by catenated strands of DNA) and a G2/M-phase block characteristic of the decatenation checkpoint. Endogenous BAF complexes interact directly with endogenous topoisomerase IIα (TOP2A) through BAF250a and are required for the binding of TOP2A to approximately 12,000 sites across the genome. Our results demonstrate that TOP2A chromatin binding is dependent on the ATPase activity of BRG1, which is compromised in oncogenic BRG1 mutants. These studies indicate that the ability of TOP2A to prevent DNA entanglement at mitosis requires BAF complexes and suggest that this activity contributes to the role of BAF subunits as tumour suppressors.
Conflict of interest statement
The authors declare no competing interests
Figures
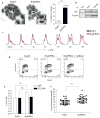
Figure 1. Brg1 associates with Topoisomerase IIα and regulates its function
a, Anaphase bridges in Brgf/f and Brgf/fERΔ ES cells. Data represent n number of slides from four independent experiments +/− SEM. b, Co-IP of Brg1 and TopoIIα from nuclear lysates. c, DNA content in Brgf/f and Brgf/fERΔ ES cells after release from double thymidine block. d, Cell cycle analysis of Brgf/f and Brgf/fERΔ ES cells with or without caffeine, an ATM/ATR inhibitor. e, Data represent 5 independent H3(S10)P cell cycle analyses +/− SEM. f, Data represent the mean of the average chromosome length per cell from 40 cells from metaphase spreads of Brgf/f and Brgf/fERΔ MEFs from 2 independent experiments.
Figure 2. Expression of Medulloblastoma-associated Brg1 mutants phenocopies Topoisomerase IIα inhibition
a, Somatic mutations in BRG1 found in medulloblastoma. b, The DNA-stimulated ATPase activity of BAF complexes from BrgWT, BrgGD, BrgKR, BrgTM, and vector control-expressing Brgf/fERΔ cells +/− SEM. c, Cell cycle analysis of Brgf/f MEFs expressing BrgWT, BrgGD, BrgTM, or vector, treated with ethanol or tamoxifen (Tax). d, Cells were prepared as in (c) and the mean frequency of anaphase bridges +/− SEM from three independent experiments was measured. e, Cells were prepared as in (c) and harvested for metaphase spreads. The number of cells with greater than 40 chromosomes (AT) was quantitated from >50 cells. Significance was calculated relative to vector control, ethanol-treated Brgf/f cells where *p<0.05, **p<0.01, ***p<0.0001. f, Various tissues were sectioned and scored for the number of anaphase bridges of total anaphases.
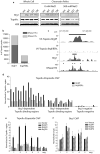
Figure 3. Brg1 facilitates the binding of Topoisomerase IIα to chromatin in vivo through ATPase-dependent chromatin remodeling activity
a, Chromatin pellets isolated from nuclei of BrgWT, BrgGD, BrgTM, and vector-expressing Brgf/fERΔ ES cells lysed in +/− 500mM NaCl. b, The number of DNase I hypersensitive TopoIIα peaks of the total number of TopoIIα peaks from TopoIIα ChIP-seqs in Brgf/f and Brgf/fERΔ ES cells. c, Representative ChIP-seq tracks for TopoIIα (in Brgf/f and Brgf/fERΔ ES cells), Brg1, and DNase I hypersensitivity. d, TopoIIα ChIP-qPCR confirmation from Brgf/f and Brgf/fERΔ ES cells. e, TopoIIα ChIP-qPCR confirmation from BrgWT, BrgGD, BrgTM, and vector control-expressing Brgf/fERΔ ES cells. f, Brg1 ChIP-qPCRs from BrgWT, BrgGD, BrgTM, and vector control-expressing Brgf/fERΔ ES cells. e, f, Data represent means of triplicate experiments +/− SEM.
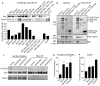
Figure 4. Topoisomerase IIα associates with the BAF complex through BAF250a
a, IPs from ES nuclear lysates. Quantitation of the precipitated TopoIIα is shown. b, V5 was precipitated from 293Ts that had been transfected with Flag-TopoIIα and either vector, V5-tagged full-length BAF250a (BAF250a FL), or V5-tagged BAF250a fragments. Lysates and anti-V5 precipitates were blotted for anti-V5, anti-Flag, and anti-Brg1. c, Brg1 was IP'ed from ES cells following BAF250a knockdown. d, MEFs with knockdown of Brg1, BAF250a or TopoIIα. Anaphase bridge frequency is calculated for seven experiments +/− SEM. Significance was calculated relative to vector control cells where *p<0.05, **p<0.01, ***p<0.0001. e, Cell cycle analysis of MEFs from (d). Data represent the mean of the % of G2/M cells normalized to vector control from four experiments +/− SEM.
Similar articles
- A chromatin remodeling complex regulates topoisomerase IIα function.
[No authors listed] [No authors listed] Cancer Discov. 2013 Jul;3(7):OF33. doi: 10.1158/2159-8290.CD-RW2013-117. Epub 2013 May 30. Cancer Discov. 2013. PMID: 23847378 - Sister chromatid decatenation: bridging the gaps in our knowledge.
Broderick R, Niedzwiedz W. Broderick R, et al. Cell Cycle. 2015;14(19):3040-4. doi: 10.1080/15384101.2015.1078039. Cell Cycle. 2015. PMID: 26266709 Free PMC article. Review. - Bloom's syndrome and PICH helicases cooperate with topoisomerase IIα in centromere disjunction before anaphase.
Rouzeau S, Cordelières FP, Buhagiar-Labarchède G, Hurbain I, Onclercq-Delic R, Gemble S, Magnaghi-Jaulin L, Jaulin C, Amor-Guéret M. Rouzeau S, et al. PLoS One. 2012;7(4):e33905. doi: 10.1371/journal.pone.0033905. Epub 2012 Apr 26. PLoS One. 2012. PMID: 22563370 Free PMC article. - RECQL5 cooperates with Topoisomerase II alpha in DNA decatenation and cell cycle progression.
Ramamoorthy M, Tadokoro T, Rybanska I, Ghosh AK, Wersto R, May A, Kulikowicz T, Sykora P, Croteau DL, Bohr VA. Ramamoorthy M, et al. Nucleic Acids Res. 2012 Feb;40(4):1621-35. doi: 10.1093/nar/gkr844. Epub 2011 Oct 19. Nucleic Acids Res. 2012. PMID: 22013166 Free PMC article. - The SWI/SNF chromatin remodelling complex: Its role in maintaining genome stability and preventing tumourigenesis.
Brownlee PM, Meisenberg C, Downs JA. Brownlee PM, et al. DNA Repair (Amst). 2015 Aug;32:127-133. doi: 10.1016/j.dnarep.2015.04.023. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25981841 Review.
Cited by
- Chromatin remodeling gene ARID2 targets cyclin D1 and cyclin E1 to suppress hepatoma cell progression.
Duan Y, Tian L, Gao Q, Liang L, Zhang W, Yang Y, Zheng Y, Pan E, Li S, Tang N. Duan Y, et al. Oncotarget. 2016 Jul 19;7(29):45863-45875. doi: 10.18632/oncotarget.10244. Oncotarget. 2016. PMID: 27351279 Free PMC article. - Human topoisomerases and their roles in genome stability and organization.
Pommier Y, Nussenzweig A, Takeda S, Austin C. Pommier Y, et al. Nat Rev Mol Cell Biol. 2022 Jun;23(6):407-427. doi: 10.1038/s41580-022-00452-3. Epub 2022 Feb 28. Nat Rev Mol Cell Biol. 2022. PMID: 35228717 Free PMC article. Review. - Taspase1 Facilitates Topoisomerase IIβ-Mediated DNA Double-Strand Breaks Driving Estrogen-Induced Transcription.
Oelschläger L, Stahl P, Kaschani F, Stauber RH, Knauer SK, Hensel A. Oelschläger L, et al. Cells. 2023 Jan 18;12(3):363. doi: 10.3390/cells12030363. Cells. 2023. PMID: 36766705 Free PMC article. - The roles of mutated SWI/SNF complexes in the initiation and development of hepatocellular carcinoma and its regulatory effect on the immune system: A review.
Hu B, Lin JZ, Yang XB, Sang XT. Hu B, et al. Cell Prolif. 2020 Apr;53(4):e12791. doi: 10.1111/cpr.12791. Epub 2020 Mar 11. Cell Prolif. 2020. PMID: 32162380 Free PMC article. Review. - Rapid chromatin repression by Aire provides precise control of immune tolerance.
Koh AS, Miller EL, Buenrostro JD, Moskowitz DM, Wang J, Greenleaf WJ, Chang HY, Crabtree GR. Koh AS, et al. Nat Immunol. 2018 Feb;19(2):162-172. doi: 10.1038/s41590-017-0032-8. Epub 2018 Jan 15. Nat Immunol. 2018. PMID: 29335648 Free PMC article.
References
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R03 DA032469/DA/NIDA NIH HHS/United States
- R01 NS046789/NS/NINDS NIH HHS/United States
- R37 NS046789/NS/NINDS NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- ImNIH/Intramural NIH HHS/United States
- R01 CA163915/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous