TETonic shift: biological roles of TET proteins in DNA demethylation and transcription - PubMed (original) (raw)
Review
TETonic shift: biological roles of TET proteins in DNA demethylation and transcription
William A Pastor et al. Nat Rev Mol Cell Biol. 2013 Jun.
Abstract
In many organisms, the methylation of cytosine in DNA has a key role in silencing 'parasitic' DNA elements, regulating transcription and establishing cellular identity. The recent discovery that ten-eleven translocation (TET) proteins are 5-methylcytosine oxidases has provided several chemically plausible pathways for the reversal of DNA methylation, thus triggering a paradigm shift in our understanding of how changes in DNA methylation are coupled to cell differentiation, embryonic development and cancer.
Figures
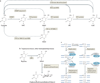
Figure 1. Mechanisms of TET-mediated demethylation
a | Known and putative pathways of DNA demethylation that involve oxidized methylcytosine intermediates. Ten-eleven translocation (TET) proteins sequentially oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). 5fC and 5caC can be removed by thymine DNA glycosylase (TDG) and replaced by cytosine via base excision repair (BER), although the extent to which this mechanism operates in specific cell types during development is unknown. Other proposed mechanisms of demethylation are less well established, including decarboxylation of 5caC, DNA methyltransferase (DNMT)-mediated removal of the hydroxymethyl group of 5hmC and deamination of 5hmC (and 5mC) (see main text) by the cytidine deaminases AID (activation-induced cytidine deaminase) and APOBEC (apolipoprotein B mRNA editing enzyme, catalytic polypeptide). AID enzymes deaminate cytosine bases in DNA to yield uracil. AID and the larger family of APOBEC enzymes have been proposed to effect DNA demethylation by deaminating 5mC and 5hmC in DNA to yield thymine and 5hmU, respectively. As these are present in mismatched T:G and 5hmU:G basepairs, they have been proposed to be excised by SMUG1 (single-strand-selective monofunctional uracil DNA glycosylase) or TDG. This mechanism is controversial, however (see main text). b | The mechanism of base J (β-
d
-glucosyl-hydroxymethyluracil) biosynthesis. The thymidine oxidation step mediated by J-binding protein 1 (JBP1) or JBP2, to produce 5-hydroxyuracil (5hmU), is analogous to the 5mC oxidation mediated by TET proteins. JBPs are the founding members of the TET–JBP superfamily: the predicted oxygenase domains of JBP1 and JBP2 were used as the starting point for the sequence profile searches that recovered the homologous domains of the three mammalian TET proteins. c | Mechanism by which 5hmC could facilitate replication-dependent DNA demethylation. A symmetrically-methylate d CpG sequence is converted during DNA replication into two asymmetrically methylated DNA strands (left panel). Hemimethylated CpG sites are recognized by UHRFI, the obligate partner of the maintenance DNA methyltransferase DNMT1, which restores symmetrical methylation. TET proteins act at methylated CpG sites to generate symmetrically hydroxymethylated CpG sequences. 5hmC and other oxizided methylcytosines may impair maintenance methylation by inhibiting UHRF1 binding, DNMT1 activity, or both (right panel). As a result, the CpG sequence progressively loses DNA methylation through successive DNA replication cycles.
Figure 2. Known protein domains of TET family members
a | Ten-eleven translocation (TET) proteins contain a DNA-binding CXXC domain towards the ami no terminus and a carboxy-terminal catalytic core region that includes a Cys-rich insert and a larger double-stranded β-helix (DSBH) domain. The number of amino acids is indicated, and the numbering corresponds to the human proteins. b | Evolutionary changes in the domain structure of TET proteins. A gene triplication event that occurred in jawed vertebrates resulted in the generation of three TET family members. A chromosomal inversion then detached the catalytic domain of TET2 from its CXXC domain, which became a separate gene (which encodes IDAX (inhibition of the Dvl and axin complex)). c | A cartoon representation of AlkB (Protein Databank (PDB) identifier: 2FD8), a protein that belongs to the same superfamily as the TET proteins and shares a common fold with them. AlkB is shown as a complex with its substrate methyladenine and its cofactor 2-oxoglutarate (2OG). In the 2OG structure, carbon atoms are shown pink, in the rest of the structure carbons are shown in grey. Nitrogens are shown in blue, oxygens in red and phosphates in orange (left panel). A stripped-down view of the active site of AlkB in complex with its substrate methlyadenine. Note the series of interactions, including pi–pi stacking interactions, between the His residues and the target base, and cation–pi interaction with the active site metal. Such interactions are likely to be retained in the TET– J-binding protein (JBP) family. In the 2OG structure, carbon atoms are shown in cyan and oxygens in red. In the protein structure, carbons are shown in grey, nitrogens in blue and oxygens in red. In methyladenine, carbons are orange and nitrogens are blue. Dashed yellow lines represent hydrogen bonds (right panel).
Figure 3. Methylation dynamics in mammalian development
a | Immediately after fertilization, the male pronucleus undergoes mass cytosine oxidation–, mediated by ten-eleventranslocation 3 (TET3). B-methylcytosine (5mC) and oxidized cytosines are then lost from the early embryo in a ‘passive’ or replication-dependent manner, resulting in the loss of nearly all modified cytosines by the 16-cell stage,. Imprinted loci retain methylation and some repetitive element classes retain partial methylation. Approximately when the blastula implants into the uterus, the inner cell mass, which gives rise to the embryo, undergoes mass de novo DNA methylation,.TET1 and TET2 are highly expressed at this stage, potentially fine-tuning methylation patterns, b | Demethylat ion also occurs in primordial germ cells (PGCs) between embryonic days E9.B and E13.B of embryonic development,,. This event also entails both mass BmC oxidation by TETl and TET2 and loss of modified cytosine by passive demethylation, resulting in the loss of imprints. A similar process of 5mC oxidation and demethylation occurs more slowly in human germ cells. This demethylat ion of imprints is critical because whereas somatic cells of an organism contain male and female imprints, the germ cells of an organism contain the imprints that correspond exclusively to the gender of the organism. Germ cells are then gradually re-methylated and imprints placed, starting at E1B in males and after birth in females.
Similar articles
- TET family dioxygenases and DNA demethylation in stem cells and cancers.
An J, Rao A, Ko M. An J, et al. Exp Mol Med. 2017 Apr 28;49(4):e323. doi: 10.1038/emm.2017.5. Exp Mol Med. 2017. PMID: 28450733 Free PMC article. Review. - Tet family proteins and 5-hydroxymethylcytosine in development and disease.
Tan L, Shi YG. Tan L, et al. Development. 2012 Jun;139(11):1895-902. doi: 10.1242/dev.070771. Development. 2012. PMID: 22569552 Free PMC article. Review. - Tet family of 5-methylcytosine dioxygenases in mammalian development.
Zhao H, Chen T. Zhao H, et al. J Hum Genet. 2013 Jul;58(7):421-7. doi: 10.1038/jhg.2013.63. Epub 2013 May 30. J Hum Genet. 2013. PMID: 23719188 Free PMC article. Review. - TET-mediated DNA demethylation controls gastrulation by regulating Lefty-Nodal signalling.
Dai HQ, Wang BA, Yang L, Chen JJ, Zhu GC, Sun ML, Ge H, Wang R, Chapman DL, Tang F, Sun X, Xu GL. Dai HQ, et al. Nature. 2016 Oct 27;538(7626):528-532. doi: 10.1038/nature20095. Epub 2016 Oct 19. Nature. 2016. PMID: 27760115 - Ten eleven translocation enzymes and 5-hydroxymethylation in mammalian development and cancer.
Kinney SR, Pradhan S. Kinney SR, et al. Adv Exp Med Biol. 2013;754:57-79. doi: 10.1007/978-1-4419-9967-2_3. Adv Exp Med Biol. 2013. PMID: 22956496 Review.
Cited by
- TET proteins regulate Drosha expression and impact microRNAs in iNKT cells.
Gioulbasani M, Äijö T, Valenzuela JE, Bettes JB, Tsagaratou A. Gioulbasani M, et al. Front Immunol. 2024 Sep 19;15:1440044. doi: 10.3389/fimmu.2024.1440044. eCollection 2024. Front Immunol. 2024. PMID: 39364402 Free PMC article. - RNA m5C oxidation by TET2 regulates chromatin state and leukaemogenesis.
Zou Z, Dou X, Li Y, Zhang Z, Wang J, Gao B, Xiao Y, Wang Y, Zhao L, Sun C, Liu Q, Yu X, Wang H, Hong J, Dai Q, Yang FC, Xu M, He C. Zou Z, et al. Nature. 2024 Oct;634(8035):986-994. doi: 10.1038/s41586-024-07969-x. Epub 2024 Oct 2. Nature. 2024. PMID: 39358506 Free PMC article. - TET1 overexpression affects cell proliferation and apoptosis in aging ovaries.
Feng Q, Li Q, Hu Y, Wang Z, Zhou H, Lin C, Wang D. Feng Q, et al. J Assist Reprod Genet. 2024 Sep 25. doi: 10.1007/s10815-024-03271-x. Online ahead of print. J Assist Reprod Genet. 2024. PMID: 39317913 - TET3 is expressed in prostate cancer tumor-associated macrophages and is associated with anti-androgen resistance.
Wei QJ, Liang HQ, Liang YW, Huang ZX. Wei QJ, et al. Clin Transl Oncol. 2024 Sep 6. doi: 10.1007/s12094-024-03708-w. Online ahead of print. Clin Transl Oncol. 2024. PMID: 39240303 - Loss of tet methyl cytosine dioxygenase 3 (TET3) enhances cardiac fibrosis via modulating the DNA damage repair response.
Rath SK, Nyamsuren G, Tampe B, Yu DS, Hulshoff MS, Schlösser D, Maamari S, Zeisberg M, Zeisberg EM. Rath SK, et al. Clin Epigenetics. 2024 Aug 27;16(1):119. doi: 10.1186/s13148-024-01719-6. Clin Epigenetics. 2024. PMID: 39192299 Free PMC article.
References
- Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET 1. Science. 2009;324:930–935.Discovery that TET proteins oxidize 5mC to 5hmC
- Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710.Describes the evolution of TET proteins and the presence of TET homologues in non-metazoan species
- Ono R, et al. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t( 10; 11] (q22;q23) Cancer Res. 2002;62:4075–4080. - PubMed
- Lorsbach RB, et al. TET1, pa member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10; 11)(q22;q23) Leukemia. 2003;17:637–641. - PubMed
- Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303.Shows that TET proteins can produce 5fC and 5caC and quantifies the level of modified cytosines in a range of cell types
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI044432/AI/NIAID NIH HHS/United States
- R01 HD065812/HD/NICHD NIH HHS/United States
- ImNIH/Intramural NIH HHS/United States
- R01 CA151535/CA/NCI NIH HHS/United States
- R01 AI44432/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources