Efficient generation of integration-free ips cells from human adult peripheral blood using BCL-XL together with Yamanaka factors - PubMed (original) (raw)
. 2013 May 21;8(5):e64496.
doi: 10.1371/journal.pone.0064496. Print 2013.
David J Baylink, Amanda Neises, Jason B Kiroyan, Xianmei Meng, Kimberly J Payne, Benjamin Tschudy-Seney, Yuyou Duan, Nancy Appleby, Mary Kearns-Jonker, Daila S Gridley, Jun Wang, K-H William Lau, Xiao-Bing Zhang
Affiliations
- PMID: 23704989
- PMCID: PMC3660366
- DOI: 10.1371/journal.pone.0064496
Efficient generation of integration-free ips cells from human adult peripheral blood using BCL-XL together with Yamanaka factors
Rui-Jun Su et al. PLoS One. 2013.
Abstract
The ability to efficiently generate integration-free induced pluripotent stem cells (iPSCs) from the most readily available source-peripheral blood-has the potential to expedite the advances of iPSC-based therapies. We have successfully generated integration-free iPSCs from cord blood (CB) CD34(+) cells with improved oriP/EBNA1-based episomal vectors (EV) using a strong spleen focus forming virus (SFFV) long terminal repeat (LTR) promoter. Here we show that Yamanaka factors (OCT4, SOX2, MYC, and KLF4)-expressing EV can also reprogram adult peripheral blood mononuclear cells (PBMNCs) into pluripotency, yet at a very low efficiency. We found that inclusion of BCL-XL increases the reprogramming efficiency by approximately 10-fold. Furthermore, culture of CD3(-)/CD19(-) cells or T/B cell-depleted MNCs for 4-6 days led to the generation of 20-30 iPSC colonies from 1 ml PB, an efficiency that is substantially higher than previously reported. PB iPSCs express pluripotency markers, form teratomas, and can be induced to differentiate in vitro into mesenchymal stem cells, cardiomyocytes, and hepatocytes. Used together, our optimized factor combination and reprogramming strategy lead to efficient generation of integration-free iPSCs from adult PB. This discovery has potential applications in iPSC banking, disease modeling and regenerative medicine.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
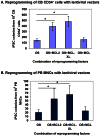
Figure 1. BCL-XL significantly enhances OS-mediated reprogramming of cord blood and peripheral blood cells with lentiviral vectors.
(A) Differential effects of BCL2 family members on enhancing OS-mediated reprogramming of CB cells. CB CD34+ cells were cultured for 2 days before lentiviral transduction. CB iPSC colonies were enumerated at 2 weeks after transduction of reprogramming factors. Data shown are presented as mean ± SEM (n = 4). OS: OCT4 and SOX2. * indicates P<0.05. (B) Differential effects of BCL2 family members on enhancing OS-mediated reprogramming of PBMNCs. PBMNCs were cultured for 4–6 days before lentiviral transduction. PB iPSC colonies were enumerated at 3 weeks after transduction of reprogramming factors. Data shown are presented as mean ± SEM (n = 4). OS: OCT4 and SOX2. PBMNCs, peripheral blood mononuclear cells. * indicates P<0.05. BCL2 and BCL-XL significantly increased reprogramming of both CB CD34+ cells and PBMNCs.
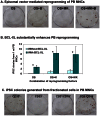
Figure 2. Generation of integration-free iPSCs from adult PBMNCs with episomal vectors.
(A) ALP staining of iPSCs at 4 weeks after nucleofection of PBMNCs with reprogramming factor-expressing episomal vectors. OS, OCT4 and SOX2; MK, MYC and KLF4; B, BCL-XL. PBMNCs were cultured for 4–8 days before nucleofection. 1×106 PBMNCs were nucleofected and then seeded into each well. (B) Inclusion of BCL-XL increases PB reprogramming efficiency by up to 10-fold. PBMNCs were cultured for 4–8 days before nucleofection. ALP-positive iPSC colonies were enumerated at 3–4 weeks after nucleofection. Data are presented as mean ± SEM (n = 6). In all 3 conditions, BCL-XL significantly increased reprogramming efficiency. * indicates P<0.05. (C) iPSC are generated from PBMNCs expressing the myeloid lineage marker, CD33, but not lymphoid cells (CD3+ and CD19+ cells) in PBMNCs. ALP staining of iPSCs at 4 weeks after nucleofection of fractionated PBMNCs with episomal vectors OS+MK+B. CD33, myeloid marker; CD3, T cell marker; CD19, B cell marker. 1×106 indicated cells were nucleofected and then seeded into each well. ALP staining was conducted at 4 weeks after nucleofection.
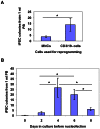
Figure 3. Optimization of the PBMNC reprogramming protocol.
(A) Depletion of CD3+ and 19+ lymphoid cells increases reprogramming efficiency. Whole PBMNCs or CD3−/CD19− cells (T/B cell-depleted PBMNCs) were cultured for 4 days before nucleofection with episomal vectors expressing OS (OCT4 and SOX2), MK (MYC and KLF4) and B (BCL-XL). 1×106 cells were used for nucleofection. The numbers of iPSC colonies were counted at 3–4 weeks after nucleofection and numbers of iPSC colonies per 1 ml of PB were calculated by normalization to the amount of starting peripheral blood. Data shown are presented as mean ± SEM (n = 4). * indicates P<0.05. (B) Culturing PB CD3/19− cells for 4 days allows maximum reprogramming. CD3/19− cells (T/B cells-depleted) PBMNCs were cultured for 0 to 8 days before nucleofection with episomal vectors expressing OS, MK and B. 1×106 cells were used for nucleofection. The numbers of iPSC colonies were counted at 3–4 weeks after nucleofection. Graphed data are presented as mean ± SEM (n = 6). * indicates P<0.05.
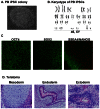
Figure 4. Characterization of integration-free PB iPSCs.
(A) PB iPSCs show a typical morphology of human pluripotent stem cells. (B) A representative karyogram of an iPSC clone. All analyzed PB iPSC clones showed a normal karyotype. (C) PB iPSCs express pluripotency markers OCT4, SOX2, NANOG and SSEA4. Shown are representative confocal images captured using the Zeiss LSM 710 confocal microscope with a 20× objective. (D) PB iPSCs form teratoma in immunodeficient mice. H & E staining of representative teratoma from PB iPSCs shows derivatives of 3 embryonic germ layers. Cartilage (mesoderm); glands (endoderm) and neurotubules (ectoderm). Images were acquired using the Olympus microscope with a 20× objective.
Figure 5. In vitro multilineage differentiation of integration-free PB iPSCs.
(A) Differentiation of PB iPSCs into MSCs. iPSC-MSCs show a typical MSC morphology and are capable of differentiation into adipocytes, osteoblasts and chondrocytes. Oil Red O stains the oil droplets of adipocytes. Alizarin Red stains the bone nodules formed by osteoblasts. Alcian Blue stains acid mucopolysaccharides synthesized and secreted by chondrocytes. (B) PB iPSC-derived hepatocytes show a typical morphology of hepatocytes at 25 days after hepatocytic differentiation. These cells also express markers AFP, albumin (ALB), and alpha 1-antitrypsin (α1-AT). The differentiated cells were stained with monoclonal antibody against AFP, goat anti-albumin, and goat anti-alpha 1-antitrypsin at 18 days after differentiation culture. (C) PB iPSC can be induced to differentiation into cardiomyocytes that express Troponin I marker. Cell nuclei were counterstained with DAPI.
Similar articles
- Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures.
Chou BK, Mali P, Huang X, Ye Z, Dowey SN, Resar LM, Zou C, Zhang YA, Tong J, Cheng L. Chou BK, et al. Cell Res. 2011 Mar;21(3):518-29. doi: 10.1038/cr.2011.12. Epub 2011 Jan 18. Cell Res. 2011. PMID: 21243013 Free PMC article. - Generation of Integration-free Induced Pluripotent Stem Cells from Human Peripheral Blood Mononuclear Cells Using Episomal Vectors.
Wen W, Zhang JP, Chen W, Arakaki C, Li X, Baylink D, Botimer GD, Xu J, Yuan W, Cheng T, Zhang XB. Wen W, et al. J Vis Exp. 2017 Jan 1;(119):55091. doi: 10.3791/55091. J Vis Exp. 2017. PMID: 28117800 Free PMC article. - Enhanced Generation of Integration-free iPSCs from Human Adult Peripheral Blood Mononuclear Cells with an Optimal Combination of Episomal Vectors.
Wen W, Zhang JP, Xu J, Su RJ, Neises A, Ji GZ, Yuan W, Cheng T, Zhang XB. Wen W, et al. Stem Cell Reports. 2016 Jun 14;6(6):873-884. doi: 10.1016/j.stemcr.2016.04.005. Epub 2016 May 5. Stem Cell Reports. 2016. PMID: 27161365 Free PMC article. - Strategies to generate induced pluripotent stem cells.
Hayes M, Zavazava N. Hayes M, et al. Methods Mol Biol. 2013;1029:77-92. doi: 10.1007/978-1-62703-478-4_6. Methods Mol Biol. 2013. PMID: 23756943 Review. - Cellular reprogramming of human peripheral blood cells.
Zhang XB. Zhang XB. Genomics Proteomics Bioinformatics. 2013 Oct;11(5):264-74. doi: 10.1016/j.gpb.2013.09.001. Epub 2013 Sep 21. Genomics Proteomics Bioinformatics. 2013. PMID: 24060839 Free PMC article. Review.
Cited by
- Episomal Induced Pluripotent Stem Cells: Functional and Potential Therapeutic Applications.
Wang AYL, Loh CYY. Wang AYL, et al. Cell Transplant. 2019 Dec;28(1_suppl):112S-131S. doi: 10.1177/0963689719886534. Epub 2019 Nov 14. Cell Transplant. 2019. PMID: 31722555 Free PMC article. Review. - Advances in Pluripotent Stem Cells: History, Mechanisms, Technologies, and Applications.
Liu G, David BT, Trawczynski M, Fessler RG. Liu G, et al. Stem Cell Rev Rep. 2020 Feb;16(1):3-32. doi: 10.1007/s12015-019-09935-x. Stem Cell Rev Rep. 2020. PMID: 31760627 Free PMC article. Review. - Positive correlation between the efficiency of induced pluripotent stem cells and the development rate of nuclear transfer embryos when the same porcine embryonic fibroblast lines are used as donor cells.
Xie B, Wang J, Liu S, Wang J, Xue B, Li J, Wei R, Zhao Y, Liu Z. Xie B, et al. Cell Reprogram. 2014 Jun;16(3):206-14. doi: 10.1089/cell.2013.0080. Epub 2014 Apr 16. Cell Reprogram. 2014. PMID: 24738969 Free PMC article. - Tissue Engineering and Regenerative Medicine 2015: A Year in Review.
Wobma H, Vunjak-Novakovic G. Wobma H, et al. Tissue Eng Part B Rev. 2016 Apr;22(2):101-13. doi: 10.1089/ten.TEB.2015.0535. Epub 2016 Feb 23. Tissue Eng Part B Rev. 2016. PMID: 26714410 Free PMC article. Review. - Reprogramming of blood cells into induced pluripotent stem cells as a new cell source for cartilage repair.
Li Y, Liu T, Van Halm-Lutterodt N, Chen J, Su Q, Hai Y. Li Y, et al. Stem Cell Res Ther. 2016 Feb 17;7:31. doi: 10.1186/s13287-016-0290-7. Stem Cell Res Ther. 2016. PMID: 26883322 Free PMC article.
References
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131: 861–872. - PubMed
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, et al. (2007) Induced pluripotent stem cell lines derived from human somatic cells. Science 318: 1917–1920. - PubMed
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, et al. (2008) Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451: 141–146. - PubMed
- Jaenisch R (2012) Nuclear cloning and direct reprogramming: the long and the short path to Stockholm. Cell Stem Cell 11: 744–747. - PubMed
- Daley GQ (2012) Cellular alchemy and the golden age of reprogramming. Cell 151: 1151–1154. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials