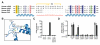Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity - PubMed (original) (raw)
Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity
Philip J Kranzusch et al. Cell Rep. 2013.
Abstract
Innate immune recognition of foreign nucleic acids induces protective interferon responses. Detection of cytosolic DNA triggers downstream immune signaling through activation of cyclic GMP-AMP synthase (cGAS). We report here the crystal structure of human cGAS, revealing an unanticipated zinc-ribbon DNA-binding domain appended to a core enzymatic nucleotidyltransferase scaffold. The catalytic core of cGAS is structurally homologous to the RNA-sensing enzyme, 2'-5' oligo-adenylate synthase (OAS), and divergent C-terminal domains account for specific ligand-activation requirements of each enzyme. We show that the cGAS zinc ribbon is essential for STING-dependent induction of the interferon response and that conserved amino acids displayed within the intervening loops are required for efficient cytosolic DNA recognition. These results demonstrate that cGAS and OAS define a family of innate immunity sensors and that structural divergence from a core nucleotidyltransferase enables second-messenger responses to distinct foreign nucleic acids.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Fig. 1. Structure of Human cGAS
(A) Cartoon schematic of the human cGAS primary sequence. (B) Overall structure of human cGAS with the N-terminal helical extension, NTase core scaffold, and carboxy-terminal domain (C-domain) shown in blue. A unique zinc-ribbon insertion is shown in magenta, and the zinc ion in yellow. (C) Structural overlay of cytosolic nucleic acid sensors, human cGAS (blue) and human OAS (pink). (D) Electrostatic surface potential of cGAS (left); a conserved positively charged nucleic-acid binding cleft equivalent to the OAS dsRNA-binding site as observed in the structure of an OAS–dsRNA complex (right, PDB code 4IG8). See also Figures S2 and S3.
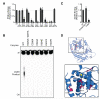
Fig. 2. In Vitro Reconstitution of cGAS Dinucleotide Signaling
(A,B) Thin layer chromatography analysis of cGAS cyclic dinucleotide synthesis. Purified full-length cGAS was incubated with substrate nucleotides and immune-stimulatory DNA (ISD DNA) as indicated. An E225A/D227A mutation to the cGAS active site (Mut) ablates cyclic dinucleotide production. Dotted radioactive spots corresponding to UV-shadowed AMP and 3′–5′ linked cGAMP markers demonstrate the product of cGAS activity is a noncanonical dinucleotide product (see also fig. S4). (C) cGAS activity is strictly dependent on dsDNA activation. (D) Fluorescence anisotropy analysis of cGAS and related Mab21L2 NTase binding to dsDNA. See also Figure S4.
Fig. 3. cGAS Zinc-Ribbon Domain Is Essential for Interferon Signaling
(A) Sequence alignment of human and murine cGAS and OAS cytosolic sensors. The unique cGAS zinc-ribbon insertion domain and coordinating residues are indicated. (B) Structural details of the zinc coordination site. Highly conserved amino acids are labeled. (C) Reconstitution of STING-dependent cGAS signaling in cells. Luciferase production under control of the interferon-β (IFNβ) promoter demonstrates that DNA-stimulated cGAS signaling only activates the interferon pathway in the presence of STING; cGAS E225A/D227A contains two point mutations in the enzymatic active site. (D) Mutational analysis of the cGAS zinc-ribbon motif. Substitutions to the zinc coordination motif (H390A, C396A, C397A, C404A) abolish cGAS activity when expressed at low levels (10 ng), and overexpression demonstrates only H390A retains weakened signaling potential (150 ng). Error bars represent the SD from the mean of at least three independent experiments.
Fig. 4. cGAS Zinc-Ribbon and Positive DNA-binding Cleft Are Essential for DNA Recognition and Catalytic Activity
(A) Reconstitution of STING-dependent cGAS signaling in cells as described in Figure 3. Single alanine and glutamine mutations to conserved positively charged amino acids within the DNA-binding cleft demonstrate K173, K384, K407 and K414 are required for efficient cytosolic DNA detection. (B) In vitro reconstitution of cGAS dinucleotide synthesis using purified components as described in Figure 2 (*P < 0.001). Mutations to the catalytic active site (E225A/D227A), zinc-coordination motif (C396A) and conserved DNA-binding cleft (K394A, K394E, K407A, K407E) all abolish DNA-stimulated enzymatic activity. (C) The ability of mutant cGAS enzymes to engage a 45 bp ISD dsDNA substrate was measured by fluorescence polarization using 2 μM of purified protein as described in Figure 2D (*P < 0.001). Mutations to the catalytic active (E225A/D227A site do no disrupt DNA interactions while disruption of the zinc-coordination motif drastically inhibits the ability of cGAS to interact with dsDNA. Error bars represent the SD from the mean of at least three independent experiments. See also Figure S1C.
Similar articles
- The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Lohöfener J, Steinke N, Kay-Fedorov P, Baruch P, Nikulin A, Tishchenko S, Manstein DJ, Fedorov R. Lohöfener J, et al. Structure. 2015 May 5;23(5):851-862. doi: 10.1016/j.str.2015.03.012. Epub 2015 Apr 16. Structure. 2015. PMID: 25892109 - Structural and functional analyses of DNA-sensing and immune activation by human cGAS.
Kato K, Ishii R, Goto E, Ishitani R, Tokunaga F, Nureki O. Kato K, et al. PLoS One. 2013 Oct 7;8(10):e76983. doi: 10.1371/journal.pone.0076983. eCollection 2013. PLoS One. 2013. PMID: 24116191 Free PMC article. - cGAS-like receptors sense RNA and control 3'2'-cGAMP signalling in Drosophila.
Slavik KM, Morehouse BR, Ragucci AE, Zhou W, Ai X, Chen Y, Li L, Wei Z, Bähre H, König M, Seifert R, Lee ASY, Cai H, Imler JL, Kranzusch PJ. Slavik KM, et al. Nature. 2021 Sep;597(7874):109-113. doi: 10.1038/s41586-021-03743-5. Epub 2021 Jul 14. Nature. 2021. PMID: 34261127 Free PMC article. - OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids.
Hornung V, Hartmann R, Ablasser A, Hopfner KP. Hornung V, et al. Nat Rev Immunol. 2014 Aug;14(8):521-8. doi: 10.1038/nri3719. Epub 2014 Jul 18. Nat Rev Immunol. 2014. PMID: 25033909 Free PMC article. Review. - Cyclic GMP-AMP as an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA.
Kato K, Omura H, Ishitani R, Nureki O. Kato K, et al. Annu Rev Biochem. 2017 Jun 20;86:541-566. doi: 10.1146/annurev-biochem-061516-044813. Epub 2017 Apr 7. Annu Rev Biochem. 2017. PMID: 28399655 Review.
Cited by
- Mechanism and therapeutic potential of targeting cGAS-STING signaling in neurological disorders.
Huang Y, Liu B, Sinha SC, Amin S, Gan L. Huang Y, et al. Mol Neurodegener. 2023 Nov 8;18(1):79. doi: 10.1186/s13024-023-00672-x. Mol Neurodegener. 2023. PMID: 37941028 Free PMC article. Review. - DNA PAMPs as Molecular Tools for the cGAS-STING Signaling Pathways.
Zheng C, Zhang L. Zheng C, et al. Methods Mol Biol. 2025;2854:117-125. doi: 10.1007/978-1-0716-4108-8_13. Methods Mol Biol. 2025. PMID: 39192124 - Structural biology of innate immunity.
Yin Q, Fu TM, Li J, Wu H. Yin Q, et al. Annu Rev Immunol. 2015;33:393-416. doi: 10.1146/annurev-immunol-032414-112258. Epub 2015 Jan 22. Annu Rev Immunol. 2015. PMID: 25622194 Free PMC article. Review. - DNA-stimulated cell death: implications for host defence, inflammatory diseases and cancer.
Paludan SR, Reinert LS, Hornung V. Paludan SR, et al. Nat Rev Immunol. 2019 Mar;19(3):141-153. doi: 10.1038/s41577-018-0117-0. Nat Rev Immunol. 2019. PMID: 30644449 Free PMC article. Review. - Control of innate immunity by the cGAS-STING pathway.
Mosallanejad K, Kagan JC. Mosallanejad K, et al. Immunol Cell Biol. 2022 Jul;100(6):409-423. doi: 10.1111/imcb.12555. Epub 2022 May 25. Immunol Cell Biol. 2022. PMID: 35485309 Free PMC article. Review.
References
- Baglioni C, Minks MA, Maroney PA. Interferon action may be mediated by activation of a nuclease by pppA2′p5′A2′p5′A. Nature. 1978;273:684–687. - PubMed
- Burdette DL, Vance RE. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol. 2013;14:19–26. - PubMed
- Daugherty MD, Malik HS. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 2012;46:677–700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials