Validation of cryo-EM structure of IP₃R1 channel - PubMed (original) (raw)
Validation of cryo-EM structure of IP₃R1 channel
Stephen C Murray et al. Structure. 2013.
Abstract
About a decade ago, three electron cryomicroscopy (cryo-EM) single-particle reconstructions of IP3R1 were reported at low resolution. It was disturbing that these structures bore little similarity to one another, even at the level of quaternary structure. Recently, we published an improved structure of IP3R1 at ∼1 nm resolution. However, this structure did not bear any resemblance to any of the three previously published structures, leading to the question of why the structure should be considered more reliable than the original three. Here, we apply several methods, including class-average/map comparisons, tilt-pair validation, and use of multiple refinement software packages, to give strong evidence for the reliability of our recent structure. The map resolution and feature resolvability are assessed with the gold standard criterion. This approach is generally applicable to assessing the validity of cryo-EM maps of other molecular machines.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
Figure 1. Class-average self-consistency test
Projections of the 3D map (A), reference-based class-averages (B), reference-free class-averages (unaligned, C), and selected (unaligned) individual particle images (D) are shown for comparison. Only a representative subset of the full set of projection orientations is shown, but clear qualitative agreement between the four types of images can be observed in each row.
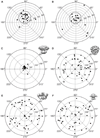
Figure 2. Results of tilt-pair analysis
(A, B) IP3R1 tilt-pair validation plots for two image tilt-pairs. The grey circles denote particle pairs that cluster around the experimental tilt geometry, thus validating our IP3R1 map (EMDB-5278)(Ludtke et al., 2011). A cross indicates the center of the cluster and each point represents a single pair of particles. The radial value indicates the amount of tilt determined between the pair of particles, and azimuthal value indicates the direction of tilt. Ideally all points would fall at exactly the experimental tilt/direction. Some spread indicates the relative uncertainty in orientation determination. Note that the radial axis extends only to 40 degrees in these plots. Corresponding statistics for used tilt-pair images is given in Table S1. (C) The same plot as in (A) is shown at larger scale. Validation plots for the same tilt-pair images as in (A) were calculated against different 3D maps, (D) EMDB-1061 (Sato et al., 2004), (E) from (Serysheva et al., 2003), and (F) from (Jiang et al., 2002). Note that the three previously published maps produce a completely random distribution with no clustering. See also Table S1 and Figure S2.
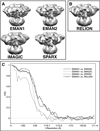
Figure 3. 3D reconstructions of IP3R1 generated with different software packages
Maps were calculated using the same set of cryo-EM images and a matching filter was applied to bring each to the same ~20 Å resolution, with a comparable contour level. (A) 3D reconstructions performed reference-free; (B) 3D structure reconstructed using an initial model generated from an EMAN2 structure that had been low pass filtered by RELION; (C) FSCs between the reconstructions from EMAN1 and the maps generated using other software packages. See also Figure S1 and Figure S2.
Figure 4. ‘Gold standard’ FSC plot for the EMAN2 reconstruction
The EMAN2 refinement of the full data set produced a ‘gold standard’ resolution of about 17 Å. Splitting the data into more homogeneous subsets resulted in an additional map (not shown) with a ‘gold standard’ resolution of 14.7 Å. This demonstrates that resolution is limited by structural variability for this data set. If the data quality were limiting the resolution, splitting the data would not improve the resolution.
Similar articles
- Validating maps from single particle electron cryomicroscopy.
Rosenthal PB, Rubinstein JL. Rosenthal PB, et al. Curr Opin Struct Biol. 2015 Oct;34:135-44. doi: 10.1016/j.sbi.2015.07.002. Epub 2015 Nov 19. Curr Opin Struct Biol. 2015. PMID: 26605834 Review. - Structural interpretation of cryo-EM image reconstructions.
Beckers M, Mann D, Sachse C. Beckers M, et al. Prog Biophys Mol Biol. 2021 Mar;160:26-36. doi: 10.1016/j.pbiomolbio.2020.07.004. Epub 2020 Jul 29. Prog Biophys Mol Biol. 2021. PMID: 32735944 - Cryo-EM of macromolecular assemblies at near-atomic resolution.
Baker ML, Zhang J, Ludtke SJ, Chiu W. Baker ML, et al. Nat Protoc. 2010 Sep;5(10):1697-708. doi: 10.1038/nprot.2010.126. Epub 2010 Sep 30. Nat Protoc. 2010. PMID: 20885381 Free PMC article. - Flexible architecture of IP3R1 by Cryo-EM.
Ludtke SJ, Tran TP, Ngo QT, Moiseenkova-Bell VY, Chiu W, Serysheva II. Ludtke SJ, et al. Structure. 2011 Aug 10;19(8):1192-9. doi: 10.1016/j.str.2011.05.003. Structure. 2011. PMID: 21827954 Free PMC article. - Refinement of Atomic Structures Against cryo-EM Maps.
Murshudov GN. Murshudov GN. Methods Enzymol. 2016;579:277-305. doi: 10.1016/bs.mie.2016.05.033. Epub 2016 Jun 24. Methods Enzymol. 2016. PMID: 27572731 Review.
Cited by
- Identifying and Visualizing Macromolecular Flexibility in Structural Biology.
Palamini M, Canciani A, Forneris F. Palamini M, et al. Front Mol Biosci. 2016 Sep 9;3:47. doi: 10.3389/fmolb.2016.00047. eCollection 2016. Front Mol Biosci. 2016. PMID: 27668215 Free PMC article. Review. - Direct visualization of DNA baton pass between replication factors bound to PCNA.
Mayanagi K, Ishino S, Shirai T, Oyama T, Kiyonari S, Kohda D, Morikawa K, Ishino Y. Mayanagi K, et al. Sci Rep. 2018 Nov 1;8(1):16209. doi: 10.1038/s41598-018-34176-2. Sci Rep. 2018. PMID: 30385773 Free PMC article. - A structural model of the genome packaging process in a membrane-containing double stranded DNA virus.
Hong C, Oksanen HM, Liu X, Jakana J, Bamford DH, Chiu W. Hong C, et al. PLoS Biol. 2014 Dec 16;12(12):e1002024. doi: 10.1371/journal.pbio.1002024. eCollection 2014 Dec. PLoS Biol. 2014. PMID: 25514469 Free PMC article. - Structure of the AcrAB-TolC multidrug efflux pump.
Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, Venter H, Chiu W, Luisi BF. Du D, et al. Nature. 2014 May 22;509(7501):512-5. doi: 10.1038/nature13205. Epub 2014 Apr 20. Nature. 2014. PMID: 24747401 Free PMC article. - Understanding IP3R channels: From structural underpinnings to ligand-dependent conformational landscape.
Baker MR, Fan G, Arige V, Yule DI, Serysheva II. Baker MR, et al. Cell Calcium. 2023 Sep;114:102770. doi: 10.1016/j.ceca.2023.102770. Epub 2023 Jun 22. Cell Calcium. 2023. PMID: 37393815 Free PMC article. Review.
References
- Chen DH, Song JL, Chuang DT, Chiu W, Ludtke SJ. An expanded conformation of single-ring GroEL-GroES complex encapsulates an 86 kDa substrate. Structure. 2006;14:1711–1722. - PubMed
- Hamada K, Terauchi A, Mikoshiba K. Three-dimensional rearrangements within inositol 1, 4, 5-trisphosphate receptor by calcium. J Biol Chem. 2003;278:52881–52889. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R21 AR063255/AR/NIAMS NIH HHS/United States
- R01GM072804/GM/NIGMS NIH HHS/United States
- R01 GM072804/GM/NIGMS NIH HHS/United States
- P41 RR002250/RR/NCRR NIH HHS/United States
- R01GM079429/GM/NIGMS NIH HHS/United States
- R21AR063255/AR/NIAMS NIH HHS/United States
- R01 GM080139/GM/NIGMS NIH HHS/United States
- R01GM080139/GM/NIGMS NIH HHS/United States
- P41GM103832/GM/NIGMS NIH HHS/United States
- P41 GM103832/GM/NIGMS NIH HHS/United States
- R01 GM079429/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources