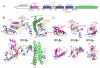Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism - PubMed (original) (raw)
Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism
David M Duda et al. Structure. 2013.
Abstract
A distinct mechanism for ubiquitin (Ub) ligation has recently been proposed for the RING1-IBR-RING2 (RBR) family of E3s: an N-terminal RING1 domain recruits a thioester-linked intermediate complex between Ub and the E2 UbcH7, and a structurally distinct C-terminal RING2 domain displays a catalytic cysteine required for Ub ligation. To obtain insights into RBR E3s, we determined the crystal structure of the human homolog of Ariadne (HHARI), which reveals the individual RING1, IBR, and RING2 domains embedded in superdomains involving sequences specific to the Ariadne RBR subfamily. The central IBR is flanked on one side by RING1, which is exposed and binds UbcH7. On the other side, a C-terminal autoinhibitory "Ariadne domain" masks the RING2 active site. Insights into RBR E3 mechanisms are provided by structure-based mutations that indicate distinct steps of relief from autoinhibition, Ub transfer from E2 to HHARI, and ligation from the HHARI cysteine to a terminal acceptor.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures
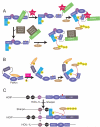
Figure 1. HHARI is inactive for autoubiquitination but unlike Parkin binds UbcH7
a. SYPRO-Ruby-stained reducing SDS-PAGE gel after time-course incubating full-length HHARI or Parkin with E1 (Uba1), UbcH7, Ub, and MgATP. b. Gel filtration chromatography profiles of Parkin or HHARI alone (green), UbcH7 (red), and of Parkin or HHARI mixed with UbcH7 (blue). SYPRO-Ruby-stained SDS-PAGE gels of fractions from the gel filtration peaks are shown below chromatograms. For reference to relative migration during chromatography, the same data with UbcH7 are shown below both chromatograms.
Figure 2. Structure of HHARI (supported by Supplementary Figure 1)
a. Schematic view of domain structure of HHARI. Colored boxes correspond to the IBR-interacting strand (IIS), UBA-like, RING1, RING1-to-IBR linker, IBR, RING2, and Ariadne domains observed in the crystal structure. White boxes correspond to regions not visible in the electron density. The location of the catalytic Cys357 is indicated in orange. b. Two views, rotated 90° around the x axis, of one molecule in the asymmetric unit for HHARI determined from the P41 crystal form, with the IBR-interacting strand in maroon, the UBA-like domain in violet, the RING1, IBR, and RING2 domains in slate, the RING1-to-IBR linker in tan, and the Ariadne domain in green. Zinc atoms are shown as yellow spheres. c. Individual domains from the RBR superimposed on related structures: HHARI RING1 (slate) superimposed on a canonical RING domain as shown from c-Cbl (1FBV.PDB, pink); HHARI IBR domain (slate) superimposed on the IBR domain from HOIP (2CT7.PDB, pink); HHARI RING2 manually docked via its C-terminal zinc-binding site on the NMR structure of the isolated HHARI RING2 bound to only a single zinc atom (1WD2.PDB, pink).
Figure 3. The Ariadne domain binds RING2 to mediate autoinhibition (supported by Supplementary Figure 3)
a. The Ariadne domain (green) masks the RING2 (slate) surface containing the catalytic Cys357 (sulfhydryl shown as orange sphere). Zinc atoms are shown as yellow spheres. b. The IBR and RING2 domains are shown in surface representation, colored by conservation among Ariadne family members, with the Ariadne domain shown in green cartoon. c. Close-up view of intramolecular contacts between the Ariadne domain (green) and RING2 domain (slate, with catalytic Cys357 sulfhydryl represented as a small orange sphere, and zinc atoms as yellow spheres). d. SYPRO-Ruby stained reducing SDS-PAGE gels showing time-courses of autoubiquitination of the full-length or mutant form of HHARI with the Ariadne domain deleted (HHARIΔAri) in the presence of E1 (Uba1), E2 (UbcH7), Ub, and MgATP. e. SYPRO-Ruby stained reducing SDS-PAGE gels showing time-courses of autoubiquitination of HHARIΔAri in the presence or absence of the isolated Ariadne domain (HHARIAri) or a mutant (HHARIAri (mut)) in the structurally-observed RING2-binding surface (F430A, E431A, E503A). f. SYPRO-Ruby stained reducing SDS-PAGE gels showing time-courses of autoubiquitination of wild-type full-length HHARI or indicated mutants with alanine substitutions in the RING2-binding surface of the Ariadne domain.
Figure 4. The Ariadne domain inhibits Ub transfer from UbcH7 to HHARI (supported by Supplementary Table 1)
a. Fluorescence scans of nonreducing SDS-PAGE gels showing time-courses of the “chase” in which the thioester-linked fluorescent lysinelesss UbK0~UbcH7 intermediate was incubated with HHARIΔAri in the presence or absence of the isolated Ariadne domain (HHARIAri) or the mutant (HHARIAri(mut)) in the structurally-observed RING2-binding surface (F430A, E431A, E503A). b. Same experiment as panel a, but after samples were reduced with DTT.
Figure 5. Recruitment of UbcH7 to HHARI RING1 is required for intrinsic autoubiquitination activity (supported by Supplementary Figure 4)
a. Gel filtration chromatography profiles of HHARIΔAriadne (HHARIΔAri) alone (green), UbcH7 alone (red) or a mixture of HHARIΔAri and UbcH7 (blue). SYPRO-Ruby-stained SDS-PAGE gels of fractions from the gel filtration peaks are shown below chromatograms. b. Isothermal Titration Calorimetry data for interactions between UbcH7 and wild-type (WT) HHARI or HHARIΔAri. Upper panels show raw power data recorded during the titration experiments, and lower panels show fits of standard binding equations after integration of the raw data, using Origin (v. 7.0) software provided by MicroCal. c. Surface representation of HHARI with UbcH5a~Ubiquitin from the RNF4~UbcH5a~Ubiquitin complex (4AP4.pdb) modeled onto RING1. The positions of the catalytic cysteines are marked by orange spheres. Ile188, a mutant defective in E2 binding, is shown in red. d. Close-up view of the predicted E2-RING1 interaction showing the positions of UbcH7 Phe62 and HHARI Ile188. e. SYPRO-Ruby-stained SDS-PAGE gel showing a time course for autoubiquitination of HHARIΔAri or HHARIΔAri I188A
Figure 6. RING2 mutations that stabilize E3 ubiquitin-thioester intermediate reveal residues required for ligation reaction
a. Cartoon representation of RING2 (slate) with showing residues from the catalytic loop. RING2 zinc ions are shown as yellow spheres. b. Fluorescence scans of SDS-PAGE gels showing time-courses of the “chase” in which the thioester-linked fluorescent Ub~UbcH7 intermediate was incubated with HHARIΔAri or HHARIΔARI catalytic loop mutants N358A or H359A. Top panel is nonreducing. Bottom panel is reducing.
Figure 7. Superposition of the crystal forms shows domain variation hinging at the IBR
Superposition of the P63 crystal form (gray) with the P41 crystal form of HHARI aligned over their IBR domains to show structural variability. Arrows indicate directions of domain rotations between the P41 and P63 crystal forms. The individual domains all superimpose well as shown below.
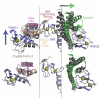
Figure 8. Models for RBR E3 ligase activation
a. Two possible models for HHARI activation. Postranslational modification (PTM) of the Ariadne domain could disrupt the interaction with RING2 and allow for RING2~Ub thioester formation and subsequent ubiquitination of substrate. Another possible activation mechanism would involve another protein binding to HHARI to release the Ariadne domain from RING2. b. Current model for Parkin activation whereby PINK1 phosphorylates the UblD domain and releases its autoinhibitory effect. c. Current model for HOIP activation where HOIL-1L and Sharpin UBL domains interact with HOIP to release autoinhibitory domains and promote ubiquitination of NEMO.
Comment in
- HHARI is one HECT of a RING.
Davies CW, Das C. Davies CW, et al. Structure. 2013 Jun 4;21(6):872-4. doi: 10.1016/j.str.2013.05.003. Structure. 2013. PMID: 23747110 Free PMC article.
Similar articles
- Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms.
Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Dove KK, et al. EMBO Rep. 2016 Aug;17(8):1221-35. doi: 10.15252/embr.201642641. Epub 2016 Jun 16. EMBO Rep. 2016. PMID: 27312108 Free PMC article. - Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Yuan L, Lv Z, Atkison JH, Olsen SK. Yuan L, et al. Nat Commun. 2017 Aug 8;8(1):211. doi: 10.1038/s41467-017-00272-6. Nat Commun. 2017. PMID: 28790309 Free PMC article. - Structural Studies of HHARI/UbcH7∼Ub Reveal Unique E2∼Ub Conformational Restriction by RBR RING1.
Dove KK, Olszewski JL, Martino L, Duda DM, Wu XS, Miller DJ, Reiter KH, Rittinger K, Schulman BA, Klevit RE. Dove KK, et al. Structure. 2017 Jun 6;25(6):890-900.e5. doi: 10.1016/j.str.2017.04.013. Epub 2017 May 25. Structure. 2017. PMID: 28552575 Free PMC article. - RING-Between-RING E3 Ligases: Emerging Themes amid the Variations.
Dove KK, Klevit RE. Dove KK, et al. J Mol Biol. 2017 Nov 10;429(22):3363-3375. doi: 10.1016/j.jmb.2017.08.008. Epub 2017 Aug 19. J Mol Biol. 2017. PMID: 28827147 Free PMC article. Review. - RBR E3-ligases at work.
Smit JJ, Sixma TK. Smit JJ, et al. EMBO Rep. 2014 Feb;15(2):142-54. doi: 10.1002/embr.201338166. Epub 2014 Jan 27. EMBO Rep. 2014. PMID: 24469331 Free PMC article. Review.
Cited by
- Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms.
Dove KK, Stieglitz B, Duncan ED, Rittinger K, Klevit RE. Dove KK, et al. EMBO Rep. 2016 Aug;17(8):1221-35. doi: 10.15252/embr.201642641. Epub 2016 Jun 16. EMBO Rep. 2016. PMID: 27312108 Free PMC article. - The role of ubiquitination in tumorigenesis and targeted drug discovery.
Deng L, Meng T, Chen L, Wei W, Wang P. Deng L, et al. Signal Transduct Target Ther. 2020 Feb 29;5(1):11. doi: 10.1038/s41392-020-0107-0. Signal Transduct Target Ther. 2020. PMID: 32296023 Free PMC article. Review. - Mechanism of phospho-ubiquitin-induced PARKIN activation.
Wauer T, Simicek M, Schubert A, Komander D. Wauer T, et al. Nature. 2015 Aug 20;524(7565):370-4. doi: 10.1038/nature14879. Epub 2015 Jul 10. Nature. 2015. PMID: 26161729 Free PMC article. - Protein clearance strategies for disease intervention.
Hommen F, Bilican S, Vilchez D. Hommen F, et al. J Neural Transm (Vienna). 2022 Feb;129(2):141-172. doi: 10.1007/s00702-021-02431-y. Epub 2021 Oct 23. J Neural Transm (Vienna). 2022. PMID: 34689261 Free PMC article. Review. - Structural and functional insights to ubiquitin-like protein conjugation.
Streich FC Jr, Lima CD. Streich FC Jr, et al. Annu Rev Biophys. 2014;43:357-79. doi: 10.1146/annurev-biophys-051013-022958. Annu Rev Biophys. 2014. PMID: 24773014 Free PMC article. Review.
References
- Ardley HC, Tan NG, Rose SA, Markham AF, Robinson PA. Features of the parkin/ariadne-like ubiquitin ligase, HHARI, that regulate its interaction with the ubiquitin-conjugating enzyme, Ubch7. The Journal of biological chemistry. 2001;276:19640–19647. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5P30 CA021765/CA/NCI NIH HHS/United States
- P41 GM103403/GM/NIGMS NIH HHS/United States
- P41 RR015301/RR/NCRR NIH HHS/United States
- R01 GM069530/GM/NIGMS NIH HHS/United States
- P30 CA021765/CA/NCI NIH HHS/United States
- 8P41 GM103403-10/GM/NIGMS NIH HHS/United States
- 5P41 RR015301-10/RR/NCRR NIH HHS/United States
- HHMI_/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases