Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome - PubMed (original) (raw)
Cell-type, allelic, and genetic signatures in the human pancreatic beta cell transcriptome
Alexandra C Nica et al. Genome Res. 2013 Sep.
Abstract
Elucidating the pathophysiology and molecular attributes of common disorders as well as developing targeted and effective treatments hinges on the study of the relevant cell type and tissues. Pancreatic beta cells within the islets of Langerhans are centrally involved in the pathogenesis of both type 1 and type 2 diabetes. Describing the differentiated state of the human beta cell has been hampered so far by technical (low resolution microarrays) and biological limitations (whole islet preparations rather than isolated beta cells). We circumvent these by deep RNA sequencing of purified beta cells from 11 individuals, presenting here the first characterization of the human beta cell transcriptome. We perform the first comparison of gene expression profiles between beta cells, whole islets, and beta cell depleted islet preparations, revealing thus beta-cell-specific expression and splicing signatures. Further, we demonstrate that genes with consistent increased expression in beta cells have neuronal-like properties, a signal previously hypothesized. Finally, we find evidence for extensive allelic imbalance in expression and uncover genetic regulatory variants (eQTLs) active in beta cells. This first molecular blueprint of the human beta cell offers biological insight into its differentiated function, including expression of key genes associated with both major types of diabetes.
Figures
Figure 1.
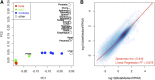
High overall similarity between beta cell and islet gene expression. (A) PCA analysis of gene RPKMs for beta (N = 11), islet (N = 7), nonbeta (N = 5) preparations, and 18 other tissues from unrelated individuals. Beta cell and islet samples cluster together, separating from nonbetas. The other tissues cluster separately, with liver being the most similar to the islet-derived RNA-seq data. (B) Scatterplot of beta cell versus islet median RPKMs on log10 scale.
Figure 2.
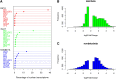
Expression differences between beta, islet, and nonbeta samples. (A) Dot chart of top 10 highest expressed genes and their contribution to the nuclear transcriptome by cell type. (B) Histogram of log2 Fold Change (islet/beta) for differentially expressed genes (10% FDR) in islets and beta cells. (C) Histogram of log2 Fold Change (nonbeta/beta) for differentially expressed genes (10% FDR) in nonbeta and beta cells.
Figure 3.
Sharing of significant ASE effects between beta cells and islets (sample P775). Left panel histograms show the enrichment (pi1) of significant ASE _P_-values in the islets for ASE sites discovered in beta cells and vice versa (beta cell ASE _P_-values of ASE sites discovered in islets). Right panel scatterplots display the direction of ASE effects (ratio of reference allele count to the total number of reads covering that site) between the two cell types, almost always in concordance.
Figure 4.
Candidate beta cell cis eQTLs discovered initially in other tissues (fat, LCL, skin). Top panel boxplots show the deviations of allelic ratios (reference/total) from the expected 0.5 in ASE individuals grouped by eQTL genotype, with heterozygotes having markedly higher effects on ASE ratios compared to homozygotes. Bottom scatterplots show the beta coefficients of the MuTHER eQTLs and the corresponding beta ASE ratios for the selected candidate beta cell regulatory variants.
Similar articles
- Genetic regulatory signatures underlying islet gene expression and type 2 diabetes.
Varshney A, Scott LJ, Welch RP, Erdos MR, Chines PS, Narisu N, Albanus RD, Orchard P, Wolford BN, Kursawe R, Vadlamudi S, Cannon ME, Didion JP, Hensley J, Kirilusha A; NISC Comparative Sequencing Program; Bonnycastle LL, Taylor DL, Watanabe R, Mohlke KL, Boehnke M, Collins FS, Parker SC, Stitzel ML. Varshney A, et al. Proc Natl Acad Sci U S A. 2017 Feb 28;114(9):2301-2306. doi: 10.1073/pnas.1621192114. Epub 2017 Feb 13. Proc Natl Acad Sci U S A. 2017. PMID: 28193859 Free PMC article. - The miRNA profile of human pancreatic islets and beta-cells and relationship to type 2 diabetes pathogenesis.
van de Bunt M, Gaulton KJ, Parts L, Moran I, Johnson PR, Lindgren CM, Ferrer J, Gloyn AL, McCarthy MI. van de Bunt M, et al. PLoS One. 2013;8(1):e55272. doi: 10.1371/journal.pone.0055272. Epub 2013 Jan 25. PLoS One. 2013. PMID: 23372846 Free PMC article. - Androgen receptor-deficient islet β-cells exhibit alteration in genetic markers of insulin secretion and inflammation. A transcriptome analysis in the male mouse.
Xu W, Niu T, Xu B, Navarro G, Schipma MJ, Mauvais-Jarvis F. Xu W, et al. J Diabetes Complications. 2017 May;31(5):787-795. doi: 10.1016/j.jdiacomp.2017.03.002. Epub 2017 Mar 9. J Diabetes Complications. 2017. PMID: 28343791 Free PMC article. - The pancreatic β-cell transcriptome and integrated-omics.
Blodgett DM, Cura AJ, Harlan DM. Blodgett DM, et al. Curr Opin Endocrinol Diabetes Obes. 2014 Apr;21(2):83-8. doi: 10.1097/MED.0000000000000051. Curr Opin Endocrinol Diabetes Obes. 2014. PMID: 24526012 Free PMC article. Review. - Genomics of Islet (Dys)function and Type 2 Diabetes.
Lawlor N, Khetan S, Ucar D, Stitzel ML. Lawlor N, et al. Trends Genet. 2017 Apr;33(4):244-255. doi: 10.1016/j.tig.2017.01.010. Epub 2017 Feb 27. Trends Genet. 2017. PMID: 28245910 Free PMC article. Review.
Cited by
- Hippo Signaling Pathway in Pancreas Development.
Wu Y, Aegerter P, Nipper M, Ramjit L, Liu J, Wang P. Wu Y, et al. Front Cell Dev Biol. 2021 May 17;9:663906. doi: 10.3389/fcell.2021.663906. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34079799 Free PMC article. Review. - Systematic Functional Characterization of Candidate Causal Genes for Type 2 Diabetes Risk Variants.
Thomsen SK, Ceroni A, van de Bunt M, Burrows C, Barrett A, Scharfmann R, Ebner D, McCarthy MI, Gloyn AL. Thomsen SK, et al. Diabetes. 2016 Dec;65(12):3805-3811. doi: 10.2337/db16-0361. Epub 2016 Aug 23. Diabetes. 2016. PMID: 27554474 Free PMC article. - Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes.
Segerstolpe Å, Palasantza A, Eliasson P, Andersson EM, Andréasson AC, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, Smith DM, Kasper M, Ämmälä C, Sandberg R. Segerstolpe Å, et al. Cell Metab. 2016 Oct 11;24(4):593-607. doi: 10.1016/j.cmet.2016.08.020. Epub 2016 Sep 22. Cell Metab. 2016. PMID: 27667667 Free PMC article. - The regulator of G-protein signaling RGS16 promotes insulin secretion and β-cell proliferation in rodent and human islets.
Vivot K, Moullé VS, Zarrouki B, Tremblay C, Mancini AD, Maachi H, Ghislain J, Poitout V. Vivot K, et al. Mol Metab. 2016 Aug 26;5(10):988-996. doi: 10.1016/j.molmet.2016.08.010. eCollection 2016 Oct. Mol Metab. 2016. PMID: 27689011 Free PMC article. - Cell Type-Selective Expression of Circular RNAs in Human Pancreatic Islets.
Kaur S, Mirza AH, Pociot F. Kaur S, et al. Noncoding RNA. 2018 Nov 27;4(4):38. doi: 10.3390/ncrna4040038. Noncoding RNA. 2018. PMID: 30486482 Free PMC article.
References
- Ahren B 2000. Autonomic regulation of islet hormone secretion—implications for health and disease. Diabetologia 43: 393–410 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources