HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis - PubMed (original) (raw)
HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis
Miao Sun et al. Proc Natl Acad Sci U S A. 2013.
Abstract
The ten-eleven translocation (TET) family of methylcytosine dioxygenases initiates demethylation of DNA and is associated with tumorigenesis in many cancers; however, the mechanism is mostly unknown. Here we identify upstream activators and downstream effectors of TET1 in breast cancer using human breast cancer cells and a genetically engineered mouse model. We show that depleting the architectural transcription factor high mobility group AT-hook 2 (HMGA2) induces TET1. TET1 binds and demethylates its own promoter and the promoter of homeobox A (HOXA) genes, enhancing its own expression and stimulating expression of HOXA genes including HOXA7 and HOXA9. Both TET1 and HOXA9 suppress breast tumor growth and metastasis in mouse xenografts. The genes comprising the HMGA2-TET1-HOXA9 pathway are coordinately regulated in breast cancer and together encompass a prognostic signature for patient survival. These results implicate the HMGA2-TET1-HOX signaling pathway in the epigenetic regulation of human breast cancer and highlight the importance of targeting methylation in specific subpopulations as a potential therapeutic strategy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
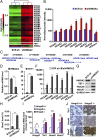
Induction of TET1 and HOX gene expression upon depletion of HMGA2 in 1833 cells, a bone-tropic derivative of human breast cancer cell line MDA-MB-231, or in MMTV_–_Wnt1 transgenic mouse breast tumors. (A, B, and D_–_H) We stably transduced 1833 cells with HMGA2 shRNA (shHMGA2) or control SCR sh. (A) Gene expression array analysis showing up-regulation of TET1 and 20 of 39 HOX genes in _HMGA2_-depleted cells. (B) The expression levels of HOXA genes are shown. *Fold change (fc) < 2; **fc > 2 based on the signal intensity of gene expression arrays. (C) Genomic transcription units of human HOXA genes on chromosome 7 viewed using the UCSC Genome Browser (40). HOXA genes are transcribed from right to left with the following order: 5′UTR (thin blue bar), Coding Sequence (thick blue bar), and 3′UTR (thin blue bar). Bar length is proportional to length of DNA sequence. (D_–_H) qRT-PCR and immunoblotting analyses validated induction of TET1 and HOXA gene expression in _HMGA2_-depleted cells. (D_–_F) HMGA2 (D), TET1 (E), or HOXA4/5/6/7/9/11 (F) mRNA analyzed by qRT-PCR (GAPDH as normalization control). (G) HMGA2, TET1, and HOXA9/7 protein analyzed by immunoblotting (GAPDH as control). (H) Genome-wide 5hmC levels analyzed by dot blot assay. (I and J) Loss of Hmga2 in MMTV_–_Wnt1 transgenic mouse breast tumors induced Tet1 and Hoxa9/7 expression. Wnt1 transgenic mice were crossed with _Hmga2_-specific knockout mice. Mouse primary breast tumors were obtained from Hmga2 wild-type (Hmga2+/+), heterozygous (Hmga2+/−), or null (H_mga2_−/−) mice. (I) Murine Hmga2, Tet1, and Hoxa9/7 mRNA analyzed by qRT-PCR (with mouse Gapdh as normalization control). (J) Murine Tet1 and Hoxa9 protein and 5hmC levels analyzed by immunostaining. (D_–_F, H, and I) Data are means ± SEM; n = 3. *P < 0.05; **P < 0.01; ***P < 0.001.
Fig. 2.
TET1 is involved in an autoregulation in human breast cancer cells. (A) TET1 binds to its own promoter. We analyzed 1833 cells expressing TET1 or control by ChIP assay with anti-TET1 or H3K4Me3 antibody followed by qPCR analysis: TET1 and H3K4Me3 binding to the CpG island proximal to the TSS of TET1 (see site-1 and -2 in
SI Materials and Methods
). Site-3 is a negative control. (B and C) HMGA2 depletion causes demethylation of CpG islands at the TET1 promoter region. We analyzed 1833 cells stably expressing shHMGA2 or SCR sh for CpG island methylation status by multiple approaches. (B) TET1 promoter region was analyzed within ±1 kb from the TSS. Methylation-specific digestions followed by qPCR distinguished between methylated CpGs vs. unmethylated or other modified (e.g., 5hmC) CpGs. The percentage of methylation vs. unmethylation (includes unmethylated or other modified C) is indicated. (C) Bisulfite sequencing of specific CpGs (see
SI Materials and Methods
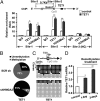
for primers) at the TET1 promoter proximal to the TSS. Results show unmethylated CpGs (open circles) vs. methylated or modified CpGs (filled circles) in 10 or more independent clones encompassing the region of interest. (D) We treated 1833 cells with 5-azacytidine followed by qRT-PCR analysis for TET1 mRNA expression (GAPDH as normalization control). (A, B, and D) Data are means ± SEM; n = 3. *P < 0.05; **P < 0.01.
Fig. 3.
TET1 induces HOXA gene expression. (A and B) Depletion of TET1 by siRNA partially countered induction of HOXA genes. We transfected 1833 cells stably expressing HMGA2 shRNA with control or TET1 siRNA. (A) Analysis of TET1 and HOXA gene mRNA by qRT-PCR. (B, Upper) Analysis of TET1 and HOXA9/7 protein by immunoblotting. (Lower) Analysis of 5hmC levels by dot-blot assay. (C and D) Expression of TET1 dramatically induced HOXA9 expression. We analyzed 1833 cells expressing constitutive Tet1 (Flag–Tet1) by qRT-PCR for HOXA9 mRNA (C) and by immunoblotting for Tet1 (Flag–M1) and HOXA9 protein (D, Upper) and dot-blot assay for 5hmC levels (D, Lower). (E and F) Induced expression of TET1 in breast xenograft tumors significantly induced HOXA9 expression. The 1833 cells stably expressing an inducible Tet1 expression vector were orthotopically injected into the mammary fat pad of nude mice. Tumor tissues were collected and analyzed after 6 wk with (+DOX) or without (−DOX) doxycycline treatment. (E) Tet1 and HOXA9 mRNA analyzed by qRT-PCR. (F) Tet1 and HOXA9 protein and 5hmC levels analyzed by immunostaining. (G) Significant positive correlation between TET1 and HOXA9/7 expression in breast cancer patients (see
Table S3
for patient information). Correlations were determined by Pearson’s correlation coefficient. P value was determined by Student t test. (A_–_E) GAPDH served as normalization control. Data are means ± SEM; n = 3. **P < 0.01; ***P < 0.001.
Fig. 4.
TET1 induces HOXA gene expression through binding to the promoter regions of HOXA genes and contributing to local demethylation in human breast cancer cells. (A and B) TET1 binds to the HOXA gene promoters. We analyzed 1833 cells expressing TET1 or control by ChIP assay with anti-TET1 or H3K4Me3 antibody followed by qPCR analysis: TET1 and H3K4Me3 binding to the CpG islands proximal to the TSS of HOXA7 (see site-1 and -2 in
SI Materials and Methods
) where site-3 is a negative control (A); or HOXA9 (see site-1 in
SI Materials and Methods
) where site-2 is a negative control (B). (C_–_E) HMGA2 depletion causes demethylation of CpG islands at HOXA gene promoter regions. We analyzed 1833 cells stably expressing shHMGA2 or SCR sh for CpG island methylation status by multiple approaches (see Fig. 2 B and C for the specificity of each method). (C) HOXA promoter regions were analyzed within −5 to +3 kb from the TSS. The percentage of methylation vs. unmethylation is indicated. (D and E) Bisulfite sequencing of specific CpGs (see
SI Materials and Methods
for primers) at HOXA7 (D) and HOXA9 (E) promoters proximal to the TSS. Results show unmethylated CpGs (open circles) vs. methylated or modified CpGs (filled circles) in 10 independent clones encompassing the region of interest. (F and G) We treated 1833 cells with 5-azacytidine followed by qRT-PCR analysis for expression of HOXA7 (F) or HOXA9 (G) mRNA (GAPDH as normalization control). (A_–_C, F, and G) Data are means ± SEM; n = 3. **P < 0.01; ***P < 0.001.
Fig. 5.
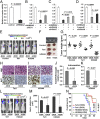
Both TET1 and its target, HOXA9, suppress breast tumor growth, invasion, and metastasis. (A_–_D) HMGA2/TET1/HOXA pathway regulates breast cancer cell invasion. (A) Inhibition of cell invasion in 1833 cells with HMGA2 depletion. (B) Transfection of TET1 siRNA into _HMGA2_-depleted 1833 cells increases invasion. (C) Transfection of HOXA7 or HOXA9 siRNA into _HMGA2_-depleted 1833 cells increases invasion. (D) Decitabine (5-aza-dC) treatment of 1833 cells decreases cell invasion, and transfection of HOXA9 siRNA into treated cells partially reversed cell invasion. (A_–_D) Data are means ± SEM; n = 3. (E_–_K) We orthotopically injected 1833 cells stably expressing an inducible control, Tet1, or HOXA9 expression vector into the mammary fat pad of nude mice. Mice were administered drinking water with (+DOX) or without (−DOX) addition of doxycycline. (E_–_G) Both TET1 and HOXA9 suppress xenograft breast tumor growth. (E) Representative bioluminescence images of mice bearing 1833 cells treated as indicated. (F) Photograph of representative xenograft breast tumors of 1833 cells treated as indicated. (G) Xenograft breast tumors of 1833 cells treated as indicated and analyzed for tumor weight. (F and G) Tumors were dissected at 6 wk after implantation. (H and I) Both TET1 and HOXA9 suppress the proliferation in xenograft breast tumors: immunostaining showing Ki67-positive cells in tumor sample of 1833 cells with induced (+DOX) vs. noninduced (−DOX) expression of Tet1 (H) or HOXA9 (I). (J and K) Both TET1 and HOXA9 inhibit intravasation of 1833 cells. Cells isolated from the blood after 6 wk were analyzed for GAPDH/Gapdh transcripts derived from human (tumor) or mouse (control) by qRT-PCR: intravasation of 1833 cells with induced (+DOX) vs. noninduced (−DOX) expression of Tet1 (J) or HOXA9 (K). Data are means ± SEM; n = 8 per group. (L_–_N) Both TET1 and HOXA9 suppress bone metastasis of 1833 cells. We injected 1833 cells stably expressing an inducible Tet1 or HOXA9 expression vector into the left ventricle of mice. Mice were administered drinking water with (+DOX) or without (−DOX) addition of doxycycline and imaged for luciferase activity after 3 wk. (L) Representative bioluminescence images of mice with bone metastasis. (M) Quantification of bone colonization by 1833 cells with induced (+DOX) vs. noninduced (−DOX) expression of Tet1 or HOXA9. Data are means ± SEM; n = 7–9 per group. (N) Kaplan–Meier survival analysis of mice over 8 wk after injection of the tumor cells.
Fig. 6.
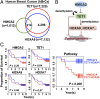
The HMGA2/TET1/HOXA pathway regulates breast tumorigenesis. (A) Comparison of the genes regulated by HMGA2, TET1, or HOXA9 in 1833 cells (human breast cancer cells; hBrCa). (B) Scheme illustrating HMGA2/TET1/HOXA signaling pathway in breast tumorigenesis. (C) Kaplan–Meier analysis of gene expression data from 101 breast tumor patients (see
Table S3
for patient information). Patients were stratified for survival using HMGA2, TET1, HOXA9, HOXA7, or the complete pathway as indicated. (Right) Red line, high HMGA2 and low TET1/HOXAs (n = 34); blue line, low HMGA2 and high TET1/HOXAs (n = 35); P, χ2 P value.
Similar articles
- TET1 upregulation drives cancer cell growth through aberrant enhancer hydroxymethylation of HMGA2 in hepatocellular carcinoma.
Shirai K, Nagae G, Seki M, Kudo Y, Kamio A, Hayashi A, Okabe A, Ota S, Tsutsumi S, Fujita T, Yamamoto S, Nakaki R, Kanki Y, Osawa T, Midorikawa Y, Tateishi K, Ichinose M, Aburatani H. Shirai K, et al. Cancer Sci. 2021 Jul;112(7):2855-2869. doi: 10.1111/cas.14897. Epub 2021 May 10. Cancer Sci. 2021. PMID: 33970549 Free PMC article. - TET1 plays an essential oncogenic role in MLL-rearranged leukemia.
Huang H, Jiang X, Li Z, Li Y, Song CX, He C, Sun M, Chen P, Gurbuxani S, Wang J, Hong GM, Elkahloun AG, Arnovitz S, Wang J, Szulwach K, Lin L, Street C, Wunderlich M, Dawlaty M, Neilly MB, Jaenisch R, Yang FC, Mulloy JC, Jin P, Liu PP, Rowley JD, Xu M, He C, Chen J. Huang H, et al. Proc Natl Acad Sci U S A. 2013 Jul 16;110(29):11994-9. doi: 10.1073/pnas.1310656110. Epub 2013 Jul 1. Proc Natl Acad Sci U S A. 2013. PMID: 23818607 Free PMC article. - TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway.
Neri F, Dettori D, Incarnato D, Krepelova A, Rapelli S, Maldotti M, Parlato C, Paliogiannis P, Oliviero S. Neri F, et al. Oncogene. 2015 Aug 6;34(32):4168-76. doi: 10.1038/onc.2014.356. Epub 2014 Nov 3. Oncogene. 2015. PMID: 25362856 - The emerging role and mechanism of HMGA2 in breast cancer.
Ma Q, Ye S, Liu H, Zhao Y, Zhang W. Ma Q, et al. J Cancer Res Clin Oncol. 2024 May 16;150(5):259. doi: 10.1007/s00432-024-05785-4. J Cancer Res Clin Oncol. 2024. PMID: 38753081 Free PMC article. Review. - Roles of the HOX Proteins in Cancer Invasion and Metastasis.
Paço A, Aparecida de Bessa Garcia S, Leitão Castro J, Costa-Pinto AR, Freitas R. Paço A, et al. Cancers (Basel). 2020 Dec 22;13(1):10. doi: 10.3390/cancers13010010. Cancers (Basel). 2020. PMID: 33375038 Free PMC article. Review.
Cited by
- HOXA9 and CD163 potentiate pancreatic ductal adenocarcinoma progression.
Hemida AS, Ahmed MM, Tantawy MS. Hemida AS, et al. Diagn Pathol. 2024 Oct 26;19(1):141. doi: 10.1186/s13000-024-01563-5. Diagn Pathol. 2024. PMID: 39462379 Free PMC article. - Active DNA Demethylase, TET1, Increases Oxidative Phosphorylation and Sensitizes Ovarian Cancer Stem Cells to Mitochondrial Complex I Inhibitor.
Chen LY, Shen YA, Chu LH, Su PH, Wang HC, Weng YC, Lin SF, Wen KC, Liew PL, Lai HC. Chen LY, et al. Antioxidants (Basel). 2024 Jun 17;13(6):735. doi: 10.3390/antiox13060735. Antioxidants (Basel). 2024. PMID: 38929174 Free PMC article. - miR-506 inhibits cell proliferation and invasion by targeting TET family in colorectal cancer.
Wu M, Zhang Y, Tang A, Tian L. Wu M, et al. Iran J Basic Med Sci. 2016 Mar;19(3):316-22. Iran J Basic Med Sci. 2016. PMID: 27114802 Free PMC article. - Peptide Sequence Influence on the Conformational Dynamics and DNA binding of the Intrinsically Disordered AT-Hook 3 Peptide.
Garabedian A, Bolufer A, Leng F, Fernandez-Lima F. Garabedian A, et al. Sci Rep. 2018 Jul 17;8(1):10783. doi: 10.1038/s41598-018-28956-z. Sci Rep. 2018. PMID: 30018295 Free PMC article. - RKIP: A Key Regulator in Tumor Metastasis Initiation and Resistance to Apoptosis: Therapeutic Targeting and Impact.
Zaravinos A, Bonavida B, Chatzaki E, Baritaki S. Zaravinos A, et al. Cancers (Basel). 2018 Aug 24;10(9):287. doi: 10.3390/cancers10090287. Cancers (Basel). 2018. PMID: 30149591 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- P50 CA125183/CA/NCI NIH HHS/United States
- CA125183-05/CA/NCI NIH HHS/United States
- CA127277/CA/NCI NIH HHS/United States
- GM087630/GM/NIGMS NIH HHS/United States
- R01 GM071440/GM/NIGMS NIH HHS/United States
- R01 CA127277/CA/NCI NIH HHS/United States
- R01 GM087630/GM/NIGMS NIH HHS/United States
- GM071440/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials