An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering - PubMed (original) (raw)
An adenosine-mediated signaling pathway suppresses prenylation of the GTPase Rap1B and promotes cell scattering
Elizabeth Ntantie et al. Sci Signal. 2013.
Abstract
During metastasis, cancer cells acquire the ability to dissociate from each other and migrate, which is recapitulated in vitro as cell scattering. The small guanosine triphosphatase (GTPase) Rap1 opposes cell scattering by promoting cell-cell adhesion, a function that requires its prenylation, or posttranslational modification with a carboxyl-terminal isoprenoid moiety, to enable its localization at cell membranes. Thus, signaling cascades that regulate the prenylation of Rap1 offer a mechanism to control the membrane localization of Rap1. We identified a signaling cascade initiated by adenosine A2B receptors that suppressed the prenylation of Rap1B through phosphorylation of Rap1B, which decreased its interaction with the chaperone protein SmgGDS (small GTPase guanosine diphosphate dissociation stimulator). These events promoted the cytosolic and nuclear accumulation of nonprenylated Rap1B and diminished cell-cell adhesion, resulting in cell scattering. We found that nonprenylated Rap1 was more abundant in mammary tumors than in normal mammary tissue in rats and that activation of adenosine receptors delayed Rap1B prenylation in breast, lung, and pancreatic cancer cell lines. Our findings support a model in which high concentrations of extracellular adenosine, such as those that arise in the tumor microenvironment, can chronically activate A2B receptors to suppress Rap1B prenylation and signaling at the cell membrane, resulting in reduced cell-cell contact and promoting cell scattering. Inhibiting A2B receptors may be an effective method to prevent metastasis.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures
Fig. 1
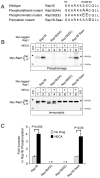
Activation of adenosine receptors induces phosphorylation of Rap1B. (A) C-terminal sequences of Rap1B phosphorylation and prenylation mutants are shown. (B) Phosphorylation of Rap1B is detectable in a phosphoimage (top) that was generated from an immunoblot (bottom) of myc-tagged Rap1B proteins that were immunoprecipitated from 32P-labeled HEK293T cells treated with or without NECA. The immunoblot (bottom) indicates different migration rates of the immunoprecipitated myc-Rap1B proteins. Asterisks indicate migration of molecular weight markers (np = non-prenylated Rap1B, p = prenylated Rap1B). (C) The fold increase in Rap1B phosphorylation was determined by measuring the O.D. of proteins detected in the phosphoimages generated in experiments described in B. The values are presented as the fold increase in relation to the O.D. of the indicated proteins from untreated cells, and are the mean +/- SEM from three independent experiments. Student’s t-test was used for statistical analysis. The asterisks indicate that phosphorylated forms of myc-tagged Rap1B(AA) and Rap1B(EE) proteins were not detected in the phosphoimages.
Fig. 2
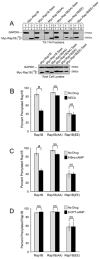
The prenylation of Rap1B is diminished by the phosphomimetic mutation and by treatment with NECA or 6-Bnz-cAMP. (A) HEK293T cells transiently expressing different myc-tagged Rap1B proteins were subjected to TX-114 fractionation to generate an aqueous phase “A” that contains non-prenylated (np) proteins, and a detergent phase “D” that contains prenylated (p) proteins. Cell fractions were immunoblotted with a myc antibody to detect the myc-tagged Rap1B proteins and a GAPDH antibody to confirm equal protein loading. An immunoblot of total cell lysates is shown beneath the TX-114 fractions. Results are representative of three independent experiments. (B-D) HEK293T cells transiently expressing different myc-tagged Rap1B proteins were treated for 24 hours with or without NECA (10 μM) (B), 6-Bnz-cAMP (1 mM) (C), or 8-CPT-cAMP (30 μM) (D). The cells were subjected to TX-114 fractionation and immunoblotting as in A. The O.D. values of the immunoblotted proteins from the aqueous and detergent phases were used to calculate the percentage of prenylated Rap1B. Values are the mean +/- SEM calculated from four (B), five (C), and three (D) independent experiments. Statistical significance was determined by matched pairs Student’s t-test with Bonferroni-adjusted p values (*, p < 0.05). Representative immunoblots are shown in Figure S2.
Fig. 3
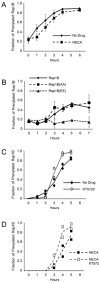
Rap1B prenylation is slowed by the phosphomimetic mutation or treatment with NECA, but accelerated by treatment with KT5720. Untransfected HEK293T cells (A) or HEK293T cells expressing myc-tagged Rap1B proteins (B) were incubated with mevastatin, washed, and placed in media with or without NECA. The cells were subjected to TX-114 fractionation at the indicated times, followed by immunoblotting using Rap1B antibody to detect endogenous Rap1B (A) or myc antibody to detect myc-tagged Rap1B proteins (B). Densitometry of the proteins in the immunoblotted aqueous and detergent fractions was performed to calculate the fraction of prenylated Rap1B. In each graph, the results are the mean +/- SEM of densitometric analysis of three independent experiments. (C, D) HEK293T cells were incubated with mevastatin, washed, incubated with or without KT5720 or NECA for the indicated times, lysed, and immunoblotted with Rap1B antibody. The fraction of prenylated Rap1B was calculated from densitometric analysis of the slowly migrating non-prenylated Rap1B and the faster migrating prenylated Rap1B detected in the immunoblots. In each graph, the results are the densitometric analysis of two independent experiments. Representative immunoblots are shown in Figure S4.
Fig. 4
Phosphorylation of Rap1B diminishes its interaction with SmgGDS-607. (A) HA immunoprecipitates from HEK293T cells coexpressing HA-tagged SmgGDS-607 with myc-tagged Rap1B or Rap1B-Saax and total cell lysates were immunoblotted with HA and myc antibodies. (B) Similar to A, but the cells were transfected with cDNAs encoding myc-tagged phosphorylation mutants of Rap1B. (C) Reticulocyte lysates were used in in vitro transcription and translation assays to generate 35S-labelled SmgGDS-607-HA and 35S-labelled myc-tagged Rap1B or a dominant negative (DN) S17N Rap1B. The 35S-labeled proteins were incubated together in the absence or presence of 6-Bnz-cAMP. Autoradiography was performed on HA immunoprecipitates from the in vitro assays. DN-Rap1B was used because it forms a more stable complex with SmgGDS-607 than does wild-type Rap1B (15). All results are representative of at least four independent experiments.
Fig. 5
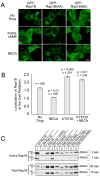
Phosphorylation of Rap1B promotes its accumulation in the cytosol and nucleus. (A) HEK293T cells expressing GFP-tagged wildtype or mutant Rap1B proteins were treated with 6-Bnz-cAMP, NECA, or no drug and imaged with confocal microscopy. All images are at the same magnification and are representative of three independent experiments. The scale bar in the lower right corner represents 10 μm. (B) HEK293T cells were transfected with a cDNA encoding GFP-Rap1B and treated with or without NECA in the presence or absence of KT5720. Images of the cells were collected by fluorescence microscopy, and the coded images were scored for membrane localization of GFP-Rap1B, as described in Figure S5. The results are the mean +/- SEM from three independent experiments. Values (n) above the columns indicate the number of scored cells. The statistical difference between the drug-treated cells compared to the untreated cells was determined by repeated-measures ANOVA with a secondary Dunnett’s multiple comparison test. (C) HEK293T cells expressing the indicated myc-tagged Rap1B proteins were lysed and the amount of GTP-bound Rap1B was measured using pulldown assays. Multiple exposures of the immunoblots are shown, as indicated by times listed at the right of the figure. Densitometric analyses of immunoblots from three independent experiments are shown in Figure S6.
Fig. 6
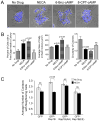
Activation of adenosine receptors promotes cell scattering, a response that is suppressed by expression of phosphodeficient Rap1B(AA). (A) Merged brightfield and DAPI images of HEK293T cells were collected after treatment with or without NECA, 6-Bnz-cAMP, or 8-CPT-cAMP. (B) Morphometric analysis of the cells described in A was conducted as explained in Figure S7. The results are the mean +/- SEM from two independent experiments with 13 – 24 colonies scored in each sample. One-way ANOVA with a secondary Dunnett’s multiple comparison test was used to determine significant differences between the drug-treated cells compared to the untreated cells. (C) HEK293T cells expressing either GFP or a GFP-tagged wild-type or mutant Rap1B protein were cultured with or without NECA before the collection of fluorescence images. The coded images were analyzed to define how many neighbors each cell contacted, as described in Figure S8. Representative images of the cells are shown in Figure S8. The results are the mean +/- SEM from two independent experiments with 18 – 21 colonies (representing 113 – 165 cells) scored in each sample. Matched-pairs Student’s t-test with Bonferonni-adjusted p values was used for statistical analysis of the bracketed samples.
Fig. 7
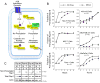
Adenosine signaling slows Rap1B prenylation in multiple cell types. (A) Our model of how adenosine regulates Rap1B. In the absence of adenosine signaling, newly synthesized Rap1B interacts with SmgGDS-607, which facilitates prenylation. Prenylated Rap1B localizes to the plasma membrane, where it promotes cell-cell adhesion. When A2BR are activated, PKA-mediated phosphorylation of Rap1B diminishes its interaction with SmgGDS-607, thereby slowing Rap1B prenylation. Non-prenylated Rap1B accumulates in the cytosol and nucleus, resulting in the loss of Rap1B functions at the plasma membrane, promoting dissolution of cell contacts and initiation of cell scattering. (B) The lung cancer lines (NCI-H1703, NCI-H23), breast cancer lines (MCF-7, MDA-MB-231) and pancreatic cancer lines (PANC-1, MiaPaCa-2) were incubated with mevastatin, washed, and incubated with or without NECA. At the indicated times, cell lysates were immunoblotted to detect endogenous Rap1B. The fraction of prenylated Rap1B was calculated from densitometric analysis of the slowly migrating non-prenylated Rap1B and the faster migrating prenylated Rap1B detected in the immunoblots. Results are the mean +/- SEM of densitometric analysis of immunoblots from 3 – 5 independent experiments (as indicated in each graph) for all cell lines except NCI-H23 cells. Results for NCI-H23 cells were obtained by densitometric analysis of immunoblots from two independent experiments. Representative immunoblots are shown in Figure S9. (C) Cell lysates prepared from mammary tumors “T” or normal mammary tissue “N” from DMBA-treated rats were immunoblotted using antibodies that detect non-prenylated Rap1, total Rap1, or actin to confirm equal loading.
Comment in
- A new signaling paradigm to control the prenylation and trafficking of small GTPases.
Williams CL. Williams CL. Cell Cycle. 2013;12(18):2933-4. doi: 10.4161/cc.26230. Epub 2013 Aug 23. Cell Cycle. 2013. PMID: 23974087 Free PMC article. No abstract available.
Similar articles
- A new signaling paradigm to control the prenylation and trafficking of small GTPases.
Williams CL. Williams CL. Cell Cycle. 2013;12(18):2933-4. doi: 10.4161/cc.26230. Epub 2013 Aug 23. Cell Cycle. 2013. PMID: 23974087 Free PMC article. No abstract available. - Adenosine promotes tumor metastasis.
Linden J. Linden J. Sci Signal. 2013 May 28;6(277):pe20. doi: 10.1126/scisignal.2004290. Sci Signal. 2013. PMID: 23716715 - Differences in the Phosphorylation-Dependent Regulation of Prenylation of Rap1A and Rap1B.
Wilson JM, Prokop JW, Lorimer E, Ntantie E, Williams CL. Wilson JM, et al. J Mol Biol. 2016 Dec 4;428(24 Pt B):4929-4945. doi: 10.1016/j.jmb.2016.10.016. Epub 2016 Oct 17. J Mol Biol. 2016. PMID: 27760305 Free PMC article. - Towards Targeting Endothelial Rap1B to Overcome Vascular Immunosuppression in Cancer.
Ghadrdoost Nakhchi B, Kosuru R, Chrzanowska M. Ghadrdoost Nakhchi B, et al. Int J Mol Sci. 2024 Sep 12;25(18):9853. doi: 10.3390/ijms25189853. Int J Mol Sci. 2024. PMID: 39337337 Free PMC article. Review. - Function, Significance, and Regulation of Rap1b in Malignancy.
Zhang L, Cui M, Song L, Zhang M, Zhang J. Zhang L, et al. Crit Rev Eukaryot Gene Expr. 2019;29(2):151-160. doi: 10.1615/CritRevEukaryotGeneExpr.2019025997. Crit Rev Eukaryot Gene Expr. 2019. PMID: 31679270 Review.
Cited by
- β-Adrenergic receptors suppress Rap1B prenylation and promote the metastatic phenotype in breast cancer cells.
Wilson JM, Lorimer E, Tyburski MD, Williams CL. Wilson JM, et al. Cancer Biol Ther. 2015;16(9):1364-74. doi: 10.1080/15384047.2015.1070988. Epub 2015 Jul 24. Cancer Biol Ther. 2015. PMID: 26209110 Free PMC article. - Long noncoding RNA LINC00514 accelerates pancreatic cancer progression by acting as a ceRNA of miR-28-5p to upregulate Rap1b expression.
Han Q, Li J, Xiong J, Song Z. Han Q, et al. J Exp Clin Cancer Res. 2020 Aug 8;39(1):151. doi: 10.1186/s13046-020-01660-5. J Exp Clin Cancer Res. 2020. PMID: 32771045 Free PMC article. - A new signaling paradigm to control the prenylation and trafficking of small GTPases.
Williams CL. Williams CL. Cell Cycle. 2013;12(18):2933-4. doi: 10.4161/cc.26230. Epub 2013 Aug 23. Cell Cycle. 2013. PMID: 23974087 Free PMC article. No abstract available. - An in vivo screening system to identify tumorigenic genes.
Ihara T, Hosokawa Y, Kumazawa K, Ishikawa K, Fujimoto J, Yamamoto M, Muramkami T, Goshima N, Ito E, Watanabe S, Semba K. Ihara T, et al. Oncogene. 2017 Apr 6;36(14):2023-2029. doi: 10.1038/onc.2016.351. Epub 2016 Oct 3. Oncogene. 2017. PMID: 27694896 - A2B Adenosine Receptor and Cancer.
Gao ZG, Jacobson KA. Gao ZG, et al. Int J Mol Sci. 2019 Oct 17;20(20):5139. doi: 10.3390/ijms20205139. Int J Mol Sci. 2019. PMID: 31627281 Free PMC article. Review.
References
- Knox AL, Brown NH. Rap1 GTPase regulation of adherens junction positioning and cell adhesion. Science. 2002;295:1285–1285. - PubMed
- Price LS, Hajdo-Milasinovic A, Zhao J, Zwartkruis FJ, Collard JG, Bos JL. Rap1 regulates E-cadherin-mediated cell-cell adhesion. J Biol Chem. 2004;20:35127–35132. - PubMed
- Yajnik V, Paulding C, Sordella R, McClatchey A, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, Settleman J, Haber DA. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–84. - PubMed
- Kooistra MRH, Dubé N, Bos JL. Rap1: a key regulator in cell-cell junction formation. J Cell Sci. 2007;120:17–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL077707/HL/NHLBI NIH HHS/United States
- R01 DK062066/DK/NIDDK NIH HHS/United States
- R01 CA125122/CA/NCI NIH HHS/United States
- R01 CA136799/CA/NCI NIH HHS/United States
- R01 CA125112/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources