Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics - PubMed (original) (raw)
Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics
Gongpu Zhao et al. Nature. 2013.
Abstract
Retroviral capsid proteins are conserved structurally but assemble into different morphologies. The mature human immunodeficiency virus-1 (HIV-1) capsid is best described by a 'fullerene cone' model, in which hexamers of the capsid protein are linked to form a hexagonal surface lattice that is closed by incorporating 12 capsid-protein pentamers. HIV-1 capsid protein contains an amino-terminal domain (NTD) comprising seven α-helices and a β-hairpin, a carboxy-terminal domain (CTD) comprising four α-helices, and a flexible linker with a 310-helix connecting the two structural domains. Structures of the capsid-protein assembly units have been determined by X-ray crystallography; however, structural information regarding the assembled capsid and the contacts between the assembly units is incomplete. Here we report the cryo-electron microscopy structure of a tubular HIV-1 capsid-protein assembly at 8 Å resolution and the three-dimensional structure of a native HIV-1 core by cryo-electron tomography. The structure of the tubular assembly shows, at the three-fold interface, a three-helix bundle with critical hydrophobic interactions. Mutagenesis studies confirm that hydrophobic residues in the centre of the three-helix bundle are crucial for capsid assembly and stability, and for viral infectivity. The cryo-electron-microscopy structures enable modelling by large-scale molecular dynamics simulation, resulting in all-atom models for the hexamer-of-hexamer and pentamer-of-hexamer elements as well as for the entire capsid. Incorporation of pentamers results in closer trimer contacts and induces acute surface curvature. The complete atomic HIV-1 capsid model provides a platform for further studies of capsid function and for targeted pharmacological intervention.
Figures
Figure 1. Cryo-EM reconstruction of HIV-1 CA tubular assembly at 8Å resolution andMDFF
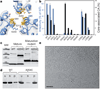
a, A cryo-EM image of recombinant A92E CA tubular assembly. Scale bar, 100 nm. b, Electron density map of the A92E CA tube with (−12, 11) helical symmetry. Yellow arrows indicate pairs of helix H9, located between adjacent hexamers. c, MDFF model of the HIV-1 capsid assembly, superimposed with the electron density map contoured at 4.0σ. Three CA hexamers, with NTDs (blue) and CTDs (orange), are shown. d,MDFF model of a CA monomer viewed from two angles. e, Two CTD dimer structures along -1 (orange) and 11 (yellow) helical directions, superimposed onto the NMR solution dimer structure (grey, 2KOD).
Figure 2. Mutational analysis of the hydrophobic trimer interface
a, Detailed viewof the trimer interface, with the structural model superimposed on the density map (grey mesh, contoured at 3.5σ). Selected residues are depicted in stick-and-ball representation. b, Virus infectivity (blue) and capsid stability (black), as percentage of total CA,± standard deviation (_n_=53), of wild-type (WT) and CA trimer interface mutants. c, Spontaneous disulphide crosslinking of A204C mature and maturation-defective virions analysed by immunoblotting for CA. d, In vitro assembly of recombinant wild-type and A204C CA proteins, assayed with high-speed sedimentation and polyacrylamide gel electrophoresis analysis. Letters u, s and p denote the unassembled reaction mixture (u) and the supernatant (s) and pellet (p) after assembly. e, Cryo-EM image of A204C in vitro assembly. Scale bar, 100 nm.
Figure 3. All-atom molecular dynamics simulation of CA tubular assembly
a, All-atom tubular assembly model comprising 71 CA hexamers (blue NTD and orange CTD) equilibrated for 125 ns. b, Ribbon representation of the tubular assembly, highlighting an HOH, circled area in a, superimposed on the density map (grey). c, Surface representation of the trimer interface (circled area in b) in a 425 ns equilibrated HOH model (hydrophobic, polar, negative and positive residues are grey, green, red and blue, respectively). d, Stereo view of a POH model after 1.5 µs equilibration. A pentamer (orange) is surrounded by five hexamers (black). e, f, Superposition of CTD H9 dimer (e) or H10 trimer (f) interfaces from HOH (blue) and POH (orange).
Figure 4. All-atom HIV-1 capsid model
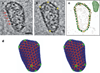
a, b, Cryo-ET analysis of an isolated, native HIV-1 core, shown as two representative slices through the three-dimensional volume. Red arrows indicate arrays of CA hexamers; yellow stars indicate locations of sharp curvature change. Scale bar, 20 nm. c, A fullerene cone model (216H+12P, green inset) matches the shape and size of the capsid, shown by the overlay of densities from the segmented capsid (orange contour) and the fullerene model (green). HIV-1 core internal densities are shown in light grey. d, Stereo view of the final molecular dynamics equilibrated all-atom capsid model (model I, see text) comprising 216 CA hexamers (blue, NTD; orange, CTD) and 12 CA pentamers (green).
Similar articles
- The structure and flexibility of conical HIV-1 capsids determined within intact virions.
Mattei S, Glass B, Hagen WJ, Kräusslich HG, Briggs JA. Mattei S, et al. Science. 2016 Dec 16;354(6318):1434-1437. doi: 10.1126/science.aah4972. Science. 2016. PMID: 27980210 - Protease cleavage leads to formation of mature trimer interface in HIV-1 capsid.
Meng X, Zhao G, Yufenyuy E, Ke D, Ning J, Delucia M, Ahn J, Gronenborn AM, Aiken C, Zhang P. Meng X, et al. PLoS Pathog. 2012;8(8):e1002886. doi: 10.1371/journal.ppat.1002886. Epub 2012 Aug 23. PLoS Pathog. 2012. PMID: 22927821 Free PMC article. - Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution.
Schur FK, Hagen WJ, Rumlová M, Ruml T, Müller B, Kräusslich HG, Briggs JA. Schur FK, et al. Nature. 2015 Jan 22;517(7535):505-8. doi: 10.1038/nature13838. Epub 2014 Nov 2. Nature. 2015. PMID: 25363765 - The capsid protein of human immunodeficiency virus: designing inhibitors of capsid assembly.
Neira JL. Neira JL. FEBS J. 2009 Nov;276(21):6110-7. doi: 10.1111/j.1742-4658.2009.07314.x. FEBS J. 2009. PMID: 19825045 Review. - The capsid protein of human immunodeficiency virus: intersubunit interactions during virus assembly.
Mateu MG. Mateu MG. FEBS J. 2009 Nov;276(21):6098-109. doi: 10.1111/j.1742-4658.2009.07313.x. FEBS J. 2009. PMID: 19825044 Review.
Cited by
- Sec24C is an HIV-1 host dependency factor crucial for virus replication.
Rebensburg SV, Wei G, Larue RC, Lindenberger J, Francis AC, Annamalai AS, Morrison J, Shkriabai N, Huang SW, KewalRamani V, Poeschla EM, Melikyan GB, Kvaratskhelia M. Rebensburg SV, et al. Nat Microbiol. 2021 Apr;6(4):435-444. doi: 10.1038/s41564-021-00868-1. Epub 2021 Mar 1. Nat Microbiol. 2021. PMID: 33649557 Free PMC article. - CryoET structures of immature HIV Gag reveal six-helix bundle.
Mendonça L, Sun D, Ning J, Liu J, Kotecha A, Olek M, Frosio T, Fu X, Himes BA, Kleinpeter AB, Freed EO, Zhou J, Aiken C, Zhang P. Mendonça L, et al. Commun Biol. 2021 Apr 16;4(1):481. doi: 10.1038/s42003-021-01999-1. Commun Biol. 2021. PMID: 33863979 Free PMC article. - Maturation of the HIV-1 core by a non-diffusional phase transition.
Frank GA, Narayan K, Bess JW Jr, Del Prete GQ, Wu X, Moran A, Hartnell LM, Earl LA, Lifson JD, Subramaniam S. Frank GA, et al. Nat Commun. 2015 Jan 8;6:5854. doi: 10.1038/ncomms6854. Nat Commun. 2015. PMID: 25569620 Free PMC article. - Microplate-based assay for identifying small molecules that bind a specific intersubunit interface within the assembled HIV-1 capsid.
Halambage UD, Wong JP, Melancon BJ, Lindsley CW, Aiken C. Halambage UD, et al. Antimicrob Agents Chemother. 2015 Sep;59(9):5190-5. doi: 10.1128/AAC.00646-15. Epub 2015 Jun 15. Antimicrob Agents Chemother. 2015. PMID: 26077250 Free PMC article. - Novel PF74-like small molecules targeting the HIV-1 capsid protein: Balance of potency and metabolic stability.
Wang L, Casey MC, Vernekar SKV, Sahani RL, Kirby KA, Du H, Zhang H, Tedbury PR, Xie J, Sarafianos SG, Wang Z. Wang L, et al. Acta Pharm Sin B. 2021 Mar;11(3):810-822. doi: 10.1016/j.apsb.2020.07.016. Epub 2020 Jul 31. Acta Pharm Sin B. 2021. PMID: 33777683 Free PMC article.
References
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283:80–83. - PubMed
- Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407:409–413. - PubMed
- Gitti RK, et al. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. - PubMed
- Momany C, et al. Crystal structure of dimeric HIV-1 capsid protein. Nature Struct. Biol. 1996;3:763–770. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases