cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING - PubMed (original) (raw)
. 2013 Jun 20;498(7454):380-4.
doi: 10.1038/nature12306. Epub 2013 May 30.
Affiliations
- PMID: 23722158
- PMCID: PMC4143541
- DOI: 10.1038/nature12306
cGAS produces a 2'-5'-linked cyclic dinucleotide second messenger that activates STING
Andrea Ablasser et al. Nature. 2013.
Abstract
Detection of cytoplasmic DNA represents one of the most fundamental mechanisms of the innate immune system to sense the presence of microbial pathogens. Moreover, erroneous detection of endogenous DNA by the same sensing mechanisms has an important pathophysiological role in certain sterile inflammatory conditions. The endoplasmic-reticulum-resident protein STING is critically required for the initiation of type I interferon signalling upon detection of cytosolic DNA of both exogenous and endogenous origin. Next to its pivotal role in DNA sensing, STING also serves as a direct receptor for the detection of cyclic dinucleotides, which function as second messenger molecules in bacteria. DNA recognition, however, is triggered in an indirect fashion that depends on a recently characterized cytoplasmic nucleotidyl transferase, termed cGAMP synthase (cGAS), which upon interaction with DNA synthesizes a dinucleotide molecule that in turn binds to and activates STING. We here show in vivo and in vitro that the cGAS-catalysed reaction product is distinct from previously characterized cyclic dinucleotides. Using a combinatorial approach based on mass spectrometry, enzymatic digestion, NMR analysis and chemical synthesis we demonstrate that cGAS produces a cyclic GMP-AMP dinucleotide, which comprises a 2'-5' and a 3'-5' phosphodiester linkage >Gp(2'-5')Ap(3'-5')>. We found that the presence of this 2'-5' linkage was required to exert potent activation of human STING. Moreover, we show that cGAS first catalyses the synthesis of a linear 2'-5'-linked dinucleotide, which is then subject to cGAS-dependent cyclization in a second step through a 3'-5' phosphodiester linkage. This 13-membered ring structure defines a novel class of second messenger molecules, extending the family of 2'-5'-linked antiviral biomolecules.
Figures
Figure 1. The R231A STING mutant uncouples cyclic di-GMP sensing from cGAS-induced activation
a, Overexpression of dinucleotide synthetases. HEK293T cells were transfected with different dinucleotide synthetases (100 ng) together with decreasing amounts of wild-type (WT) mmSTING or the R231A mutant (10, 5, 2.5, 1.25 and 0 ng) and a pIFNβ-luciferase reporter (pIFNβ-GLuc). Reporter activity was measured 16 h after transfection. RLU, relative light units. b, Direct stimulation with synthetic compounds. HEK293T cells were transfected with WT mmSTING or the R231A mutant in conjunction with pIFNβ-GLuc. The next day CMA was added or synthetic cyclic di-GMP or synthetic cGAMP(3′-5′) was transfected as indicated and pIFNβ-GLuc activity was assayed 16 h later. Representative data of two (a) or three (b) independent experiments are shown (mean values + s.e.m.).
Figure 2. The cGAS reaction product is distinct from cGAMP(3′-5′)
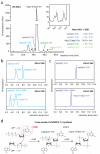
a, RP-HPLC chromatograms of lysates of untreated HEK293T cells (grey), of cGAS overexpressing HEK293T cells (light blue) or of synthetic cGAMP(3′-5′) spiked into untreated HEK293T lysate (dark blue). Asterisks highlight differential elution peaks. b, IFN-stimulated response element (ISRE) activity in LL171 cells. Endogenous cGAS product was purified from a and transfected into LL171 cells, whereas synthetic cGAMP(3′-5′) served as a control. ISRE-reporter activity was measured 14 h later. c, Chromatogram of an in vitro cGAS assay. The asterisk indicates the fraction that elutes at the same retention time as the endogenous product from a. d, ISRE activity in LL171 cells. Peaks 1-8 from c were fractionated and transfected into LL171 cells that were then studied for ISRE-reporter activity using respective control stimuli. e, TLC analysis of in-vitro- and _in-vivo_-synthesized cGAS product with ATP, GTP and synthetic cGAMP(3′-5′) as controls. f, ESI-LC-MS analysis of _in-vivo_-produced cGAS product and synthetic cGAMP(3′-5′). Representative data of two (e) or three (a-d, f) independent experiments are shown (mean values + s.e.m.).
Figure 3. The second messenger produced by cGAS is >Gp(2′-5′)Ap(3′-5′)>
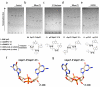
a-e, TLC analysis of GMP (lane 1), synthetic cGAMP(3′-5′) (lane 2) and enzyme-treated synthetic cGAMP(3′-5′) (lane 3), _in-vivo_-synthesized cGAS product (lane 4) and _in-vitro_-synthesized cGAS product (lane 5). Enzyme treatments of molecules analysed in lanes 3-5 were control (a), RNase T2 (b), S1 nuclease (c), RNase T1 (d) and SVPDE (e). The resulting reaction products from lanes 4 and 5 as confirmed by ESI-LC-MS analysis are depicted below. Representative data out of two independent experiments are shown. f, g, Comparison of the structure of cyclic di-GMP (4F9G.pdb) and a model for cGAMP(2′-5′) based on NMR-derived ribose conformations.
Figure 4. cGAMP(2′-5′) is a potent activator of human and murine STING
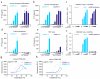
a, b, d, e, IP-10 production of murine embryonic fibroblasts, murine macrophages, human fibroblasts and THP1 cells transfected with increasing amounts of cGAMP(2′-5′) or cGAMP(3′-5′). c, f, HEK293T cells transfected with human or murine STING (0, 3.13, 6.25, 12.5 and 25 ng) as indicated, together with cGAS or DncV cGAMP synthetase (100 ng) subsequently analysed for pIFNβ-GLuc activity. g, h, Interaction of the human and murine STING LBD with 1 mM cGAMP(2′-5′) or cGAMP(3′-5′) analysed by DSF. Mean + s.e.m. of two (e) or three (a, b, d) independent experiments or one representative experiment out of three independent experiments (c, f, g, h) are depicted (c, f, mean values + s.e.m.).
Figure 5. >Gp(2′-5′)Ap(3′-5′)> is synthesized in a two-step process
a, RP-HPLC chromatogram of a cGAS+ATP+GTP in vitro reaction upon termination of the reaction. The insertion represents an enlargement of the chromatogram indicating the position of pppGp(2′-5′)G and pppGp(2′-5′)A. Right panel demonstrates mean area under the curve (AUC) + s.e.m. for depicted dinucleotides out of two independent experiments. b, c, ESI-LC-MS chromatograms before (top) and after (bottom) in vitro incubation of pppGp(2′-5′)G (b) and pppGp(2′-5′)A (c) with cGAS. Asterisks indicate the position of the substrates (*) and the resulting products (**). Note that the fraction of pppGp(2′-5′)G was contaminated with ADP. Data are representative of two independent experiments. d, Schematic model of the two-step process of cGAS-catalysed cyclic dinucleotide synthesis.
Similar articles
- Cyclic GMP-AMP containing mixed phosphodiester linkages is an endogenous high-affinity ligand for STING.
Zhang X, Shi H, Wu J, Zhang X, Sun L, Chen C, Chen ZJ. Zhang X, et al. Mol Cell. 2013 Jul 25;51(2):226-35. doi: 10.1016/j.molcel.2013.05.022. Epub 2013 Jun 6. Mol Cell. 2013. PMID: 23747010 Free PMC article. - The innate immune DNA sensor cGAS produces a noncanonical cyclic dinucleotide that activates human STING.
Diner EJ, Burdette DL, Wilson SC, Monroe KM, Kellenberger CA, Hyodo M, Hayakawa Y, Hammond MC, Vance RE. Diner EJ, et al. Cell Rep. 2013 May 30;3(5):1355-61. doi: 10.1016/j.celrep.2013.05.009. Epub 2013 May 23. Cell Rep. 2013. PMID: 23707065 Free PMC article. - Molecular mechanisms and cellular functions of cGAS-STING signalling.
Hopfner KP, Hornung V. Hopfner KP, et al. Nat Rev Mol Cell Biol. 2020 Sep;21(9):501-521. doi: 10.1038/s41580-020-0244-x. Epub 2020 May 18. Nat Rev Mol Cell Biol. 2020. PMID: 32424334 Review. - The cGAS-STING pathway for DNA sensing.
Xiao TS, Fitzgerald KA. Xiao TS, et al. Mol Cell. 2013 Jul 25;51(2):135-9. doi: 10.1016/j.molcel.2013.07.004. Mol Cell. 2013. PMID: 23870141 Free PMC article. Review. - Cyclic Guanosine Monophosphate-Adenosine Monophosphate Synthase (cGAS), a Multifaceted Platform of Intracellular DNA Sensing.
Verrier ER, Langevin C. Verrier ER, et al. Front Immunol. 2021 Feb 23;12:637399. doi: 10.3389/fimmu.2021.637399. eCollection 2021. Front Immunol. 2021. PMID: 33708225 Free PMC article. Review.
Cited by
- The Trinity of cGAS, TLR9, and ALRs Guardians of the Cellular Galaxy Against Host-Derived Self-DNA.
Kumar V. Kumar V. Front Immunol. 2021 Feb 11;11:624597. doi: 10.3389/fimmu.2020.624597. eCollection 2020. Front Immunol. 2021. PMID: 33643304 Free PMC article. Review. - Ligand-induced Ordering of the C-terminal Tail Primes STING for Phosphorylation by TBK1.
Tsuchiya Y, Jounai N, Takeshita F, Ishii KJ, Mizuguchi K. Tsuchiya Y, et al. EBioMedicine. 2016 Jul;9:87-96. doi: 10.1016/j.ebiom.2016.05.039. Epub 2016 Jun 1. EBioMedicine. 2016. PMID: 27333035 Free PMC article. - Structural basis for sequestration and autoinhibition of cGAS by chromatin.
Michalski S, de Oliveira Mann CC, Stafford CA, Witte G, Bartho J, Lammens K, Hornung V, Hopfner KP. Michalski S, et al. Nature. 2020 Nov;587(7835):678-682. doi: 10.1038/s41586-020-2748-0. Epub 2020 Sep 10. Nature. 2020. PMID: 32911480 - S6K-STING interaction regulates cytosolic DNA-mediated activation of the transcription factor IRF3.
Wang F, Alain T, Szretter KJ, Stephenson K, Pol JG, Atherton MJ, Hoang HD, Fonseca BD, Zakaria C, Chen L, Rangwala Z, Hesch A, Chan ESY, Tuinman C, Suthar MS, Jiang Z, Ashkar AA, Thomas G, Kozma SC, Gale M Jr, Fitzgerald KA, Diamond MS, Mossman K, Sonenberg N, Wan Y, Lichty BD. Wang F, et al. Nat Immunol. 2016 May;17(5):514-522. doi: 10.1038/ni.3433. Epub 2016 Apr 4. Nat Immunol. 2016. PMID: 27043414 Free PMC article. - Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape.
Hancks DC, Hartley MK, Hagan C, Clark NL, Elde NC. Hancks DC, et al. PLoS Genet. 2015 May 5;11(5):e1005203. doi: 10.1371/journal.pgen.1005203. eCollection 2015 May. PLoS Genet. 2015. PMID: 25942676 Free PMC article.
References
- Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10:123–130. - PubMed
- Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 243046/ERC_/European Research Council/International
- 322869/ERC_/European Research Council/International
- U19 AI083025/AI/NIAID NIH HHS/United States
- U19AI083025/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials